当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
CRISPR-Cas13a Based Bacterial Detection Platform: Sensing Pathogen Staphylococcus aureus in Food Samples
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2020-08-01 , DOI: 10.1016/j.aca.2020.06.041 Jin Zhou 1 , Lijuan Yin 1 , Yanan Dong 2 , Lei Peng 1 , Guozhen Liu 3 , Shuli Man 1 , Long Ma 1
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2020-08-01 , DOI: 10.1016/j.aca.2020.06.041 Jin Zhou 1 , Lijuan Yin 1 , Yanan Dong 2 , Lei Peng 1 , Guozhen Liu 3 , Shuli Man 1 , Long Ma 1
Affiliation
|
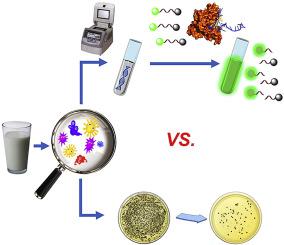
The severity of foodborne diseases caused by foods contaminated by pathogens or their toxins creates an urgent need for the development of specific and sensitive method for detection of bacteria. In this study, taking advantages of CRISPR-Cas13a system, namely, the crRNA programmability and Cas13a "collateral effect" of promiscuous RNase activity upon target RNA recognition, we developed a bacterial sensing strategy with the name of CCB-Detection (CRISPR-Cas13a based Bacterial Detection). Staphylococcus aureus (S. aureus) was chosen as a model bacteria for validating the performance of CCB-Detection. Specifically, four steps were carried out: 1) simple extraction of genome DNA; 2) specific gene amplification by PCR; 3) in vitro transcription; and 4) the "collateral effect" cleavage of reporter RNA to report the analyte signal. It was observed that CCB-Detection was capable to successfully detect the target genomic DNA (gDNA) as low as 100 aM. The limit of detection (LOD) was 1 CFU/mL with a dynamic detection range of S. aureus from 100 to 107 CFU/mL. The entire sample-to-answer time for this biosensor was less than 4 h. CCB-Detection demonstrated satisfactory selectivity for S. aureus without interference from other bacteria. Furthermore, CCB-Detection was successfully applied for sensing S. aureus in real food samples with both known and unknown amounts bacteria (spiked ones and non-spiked ones) and its performance is comparable to the conventional culture-based counting method but with short assay time and high sensitivity. With desirable reliability, sensitivity, specificity and simplicity, herein proposed CCB-Detection could be extended and generalised for other bacterial detection, and has great potential to be used in a wide range of applications such as food safety inspection, disease diagnosis, environment monitoring, etc.
中文翻译:

基于 CRISPR-Cas13a 的细菌检测平台:检测食品样品中的病原体金黄色葡萄球菌
由病原体或其毒素污染的食物引起的食源性疾病的严重性迫切需要开发特异性和灵敏的细菌检测方法。在本研究中,利用 CRISPR-Cas13a 系统的优势,即 crRNA 可编程性和 Cas13a 混杂 RNase 活性对目标 RNA 识别的“附带效应”,我们开发了一种名为 CCB-Detection (CRISPR-Cas13a based细菌检测)。选择金黄色葡萄球菌 (S. aureus) 作为模型细菌,用于验证 CCB 检测的性能。具体进行了四个步骤:1)基因组DNA的简单提取;2)PCR特异性基因扩增;3) 体外转录;和 4) 报告 RNA 的“附带效应”裂解以报告分析物信号。据观察,CCB-Detection 能够成功检测低至 100 aM 的目标基因组 DNA (gDNA)。检测限 (LOD) 为 1 CFU/mL,金黄色葡萄球菌的动态检测范围为 100 至 107 CFU/mL。该生物传感器的整个样本到回答时间不到 4 小时。CCB-Detection 对金黄色葡萄球菌表现出令人满意的选择性,而不受其他细菌的干扰。此外,CCB-Detection 成功应用于检测含有已知和未知数量细菌(掺入和非掺入细菌)的真实食品样品中的金黄色葡萄球菌,其性能可与传统的基于培养的计数方法相媲美,但检测时间短时间和高灵敏度。具有理想的可靠性、敏感性、特异性和简单性,
更新日期:2020-08-01
中文翻译:

基于 CRISPR-Cas13a 的细菌检测平台:检测食品样品中的病原体金黄色葡萄球菌
由病原体或其毒素污染的食物引起的食源性疾病的严重性迫切需要开发特异性和灵敏的细菌检测方法。在本研究中,利用 CRISPR-Cas13a 系统的优势,即 crRNA 可编程性和 Cas13a 混杂 RNase 活性对目标 RNA 识别的“附带效应”,我们开发了一种名为 CCB-Detection (CRISPR-Cas13a based细菌检测)。选择金黄色葡萄球菌 (S. aureus) 作为模型细菌,用于验证 CCB 检测的性能。具体进行了四个步骤:1)基因组DNA的简单提取;2)PCR特异性基因扩增;3) 体外转录;和 4) 报告 RNA 的“附带效应”裂解以报告分析物信号。据观察,CCB-Detection 能够成功检测低至 100 aM 的目标基因组 DNA (gDNA)。检测限 (LOD) 为 1 CFU/mL,金黄色葡萄球菌的动态检测范围为 100 至 107 CFU/mL。该生物传感器的整个样本到回答时间不到 4 小时。CCB-Detection 对金黄色葡萄球菌表现出令人满意的选择性,而不受其他细菌的干扰。此外,CCB-Detection 成功应用于检测含有已知和未知数量细菌(掺入和非掺入细菌)的真实食品样品中的金黄色葡萄球菌,其性能可与传统的基于培养的计数方法相媲美,但检测时间短时间和高灵敏度。具有理想的可靠性、敏感性、特异性和简单性,



























 京公网安备 11010802027423号
京公网安备 11010802027423号