当前位置:
X-MOL 学术
›
Metallomics
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification of novel activators of the metal responsive transcription factor (MTF-1) using a gene expression biomarker in a microarray compendium.
Metallomics ( IF 3.4 ) Pub Date : 2020-07-06 , DOI: 10.1039/d0mt00071j Abigail C Jackson 1 , Jie Liu 2 , Beena Vallanat 2 , Carlton Jones 2 , Mark D Nelms 3 , Grace Patlewicz 2 , J Christopher Corton 2
Metallomics ( IF 3.4 ) Pub Date : 2020-07-06 , DOI: 10.1039/d0mt00071j Abigail C Jackson 1 , Jie Liu 2 , Beena Vallanat 2 , Carlton Jones 2 , Mark D Nelms 3 , Grace Patlewicz 2 , J Christopher Corton 2
Affiliation
|
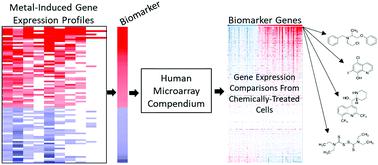
Environmental exposure to metals is known to cause a number of human toxicities including cancer. Metal-responsive transcription factor 1 (MTF-1) is an important component of metal regulation systems in mammalian cells. Here, we describe a novel method to identify chemicals that activate MTF-1 based on microarray profiling data. MTF-1 biomarker genes were identified that exhibited consistent, robust expression across 10 microarray comparisons examining the effects of metals (zinc, nickel, lead, arsenic, mercury, and silver) on gene expression in human cells. A subset of the resulting 81 biomarker genes was shown to be altered by knockdown of the MTF1 gene including metallothionein family members and a zinc transporter. The ability to correctly identify treatment conditions that activate MTF-1 was determined by comparing the biomarker to microarray comparisons from cells exposed to reference metal activators of MTF-1 using the rank-based Running Fisher algorithm. The balanced accuracy for prediction was 93%. The biomarker was then used to identify organic chemicals that activate MTF-1 from a compendium of 11 725 human gene expression comparisons representing 2582 chemicals. There were 700 chemicals identified that included those known to interact with cellular metals, such as clioquinol and disulfiram, as well as a set of novel chemicals. All nine of the novel chemicals selected for validation were confirmed to activate MTF-1 biomarker genes in MCF-7 cells and to lesser extents in MTF1-null cells by qPCR and targeted RNA-Seq. Overall, our work demonstrates that the biomarker for MTF-1 coupled with the Running Fisher test is a reliable strategy to identify novel chemical modulators of metal homeostasis using gene expression profiling.
中文翻译:

使用微阵列纲要中的基因表达生物标志物鉴定金属反应性转录因子 (MTF-1) 的新型激活剂。
众所周知,环境暴露于金属会导致多种人类毒性,包括癌症。金属反应转录因子 1 (MTF-1) 是哺乳动物细胞金属调节系统的重要组成部分。在这里,我们描述了一种新的方法来识别基于微阵列分析数据激活 MTF-1 的化学物质。鉴定出 MTF-1 生物标志物基因在 10 次微阵列比较中表现出一致、稳健的表达,以检查金属(锌、镍、铅、砷、汞和银)对人类细胞中基因表达的影响。由此产生的 81 个生物标志物基因的一个子集显示通过MTF1的敲低而改变基因包括金属硫蛋白家族成员和锌转运蛋白。正确识别激活 MTF-1 的治疗条件的能力是通过使用基于等级的 Running Fisher 算法将生物标志物与暴露于 MTF-1 参考金属激活剂的细胞的微阵列比较来确定的。预测的平衡准确度为 93%。然后使用该生物标志物从代表 2582 种化学物质的 11 725 个人类基因表达比较纲要中识别激活 MTF-1 的有机化学物质。确定了 700 种化学物质,其中包括已知与细胞金属相互作用的化学物质,如氯碘羟喹和双硫仑,以及一组新型化学物质。选择用于验证的所有九种新化学物质都被证实可激活 MCF-7 细胞中的 MTF-1 生物标志物基因,并在较小程度上激活通过 qPCR 和靶向 RNA-Seq 检测MTF1缺失细胞。总体而言,我们的工作表明,MTF-1 的生物标志物与 Running Fisher 测试相结合是使用基因表达谱鉴定金属稳态的新型化学调节剂的可靠策略。
更新日期:2020-07-06
中文翻译:

使用微阵列纲要中的基因表达生物标志物鉴定金属反应性转录因子 (MTF-1) 的新型激活剂。
众所周知,环境暴露于金属会导致多种人类毒性,包括癌症。金属反应转录因子 1 (MTF-1) 是哺乳动物细胞金属调节系统的重要组成部分。在这里,我们描述了一种新的方法来识别基于微阵列分析数据激活 MTF-1 的化学物质。鉴定出 MTF-1 生物标志物基因在 10 次微阵列比较中表现出一致、稳健的表达,以检查金属(锌、镍、铅、砷、汞和银)对人类细胞中基因表达的影响。由此产生的 81 个生物标志物基因的一个子集显示通过MTF1的敲低而改变基因包括金属硫蛋白家族成员和锌转运蛋白。正确识别激活 MTF-1 的治疗条件的能力是通过使用基于等级的 Running Fisher 算法将生物标志物与暴露于 MTF-1 参考金属激活剂的细胞的微阵列比较来确定的。预测的平衡准确度为 93%。然后使用该生物标志物从代表 2582 种化学物质的 11 725 个人类基因表达比较纲要中识别激活 MTF-1 的有机化学物质。确定了 700 种化学物质,其中包括已知与细胞金属相互作用的化学物质,如氯碘羟喹和双硫仑,以及一组新型化学物质。选择用于验证的所有九种新化学物质都被证实可激活 MCF-7 细胞中的 MTF-1 生物标志物基因,并在较小程度上激活通过 qPCR 和靶向 RNA-Seq 检测MTF1缺失细胞。总体而言,我们的工作表明,MTF-1 的生物标志物与 Running Fisher 测试相结合是使用基因表达谱鉴定金属稳态的新型化学调节剂的可靠策略。


























 京公网安备 11010802027423号
京公网安备 11010802027423号