当前位置:
X-MOL 学术
›
Immunol. Cell Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Detecting pathogenic variants in autoimmune diseases using high‐throughput sequencing
Immunology and Cell Biology ( IF 4 ) Pub Date : 2020-07-05 , DOI: 10.1111/imcb.12372 Matt A Field 1, 2
Immunology and Cell Biology ( IF 4 ) Pub Date : 2020-07-05 , DOI: 10.1111/imcb.12372 Matt A Field 1, 2
Affiliation
|
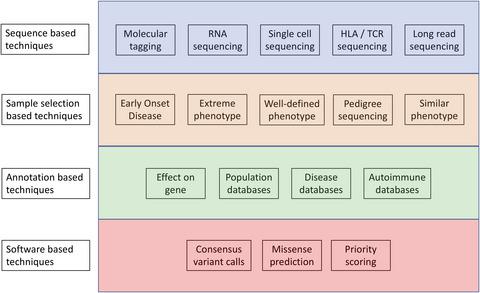
Sequencing the first human genome in 2003 took 15 years and cost $2.7 billion. Advances in sequencing technologies have since decreased costs to the point where it is now feasible to resequence a whole human genome for $1000 in a single day. These advances have allowed the generation of huge volumes of high‐quality human sequence data used to construct increasingly large catalogs of both population‐level and disease‐causing variation. The existence of such databases, coupled with a high‐quality human reference genome, means we are able to interrogate and annotate all types of genetic variation and identify pathogenic variants for many diseases. Increasingly, sequencing‐based approaches are being used to elucidate the underlying genetic cause of autoimmune diseases, a group of roughly 80 polygenic diseases characterized by abnormal immune responses where healthy tissue is attacked. Although sequence data generation has become routine and affordable, significant challenges remain with no gold‐standard methodology to identify pathogenic variants currently available. This review examines the latest methodologies used to identify pathogenic variants in autoimmune diseases and considers available sequencing options and subsequent bioinformatic methodologies and strategies. The development of reliable and robust sequencing and analytic workflows to detect pathogenic variants is critical to realize the potential of precision medicine programs where patient variant information is used to inform clinical practice.
中文翻译:

使用高通量测序检测自身免疫性疾病的致病变异
2003 年对第一个人类基因组进行测序耗时 15 年,耗资 27 亿美元。测序技术的进步已经将成本降低到现在可以在一天内以 1000 美元的价格对整个人类基因组进行重新测序的程度。这些进步使得产生了大量高质量的人类序列数据,用于构建越来越大的人口水平和致病变异的目录。此类数据库的存在,再加上高质量的人类参考基因组,意味着我们能够查询和注释所有类型的遗传变异,并确定许多疾病的致病变异。越来越多的基于测序的方法被用于阐明自身免疫性疾病的潜在遗传原因,一组大约 80 种多基因疾病,其特征是健康组织受到攻击的异常免疫反应。尽管序列数据生成已成为常规且负担得起的,但由于目前尚无确定致病变异的金标准方法,仍然存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。目前尚无确定致病变异的金标准方法,因此仍然存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。目前尚无确定致病变异的金标准方法,但仍存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于告知临床实践。
更新日期:2020-07-05
中文翻译:

使用高通量测序检测自身免疫性疾病的致病变异
2003 年对第一个人类基因组进行测序耗时 15 年,耗资 27 亿美元。测序技术的进步已经将成本降低到现在可以在一天内以 1000 美元的价格对整个人类基因组进行重新测序的程度。这些进步使得产生了大量高质量的人类序列数据,用于构建越来越大的人口水平和致病变异的目录。此类数据库的存在,再加上高质量的人类参考基因组,意味着我们能够查询和注释所有类型的遗传变异,并确定许多疾病的致病变异。越来越多的基于测序的方法被用于阐明自身免疫性疾病的潜在遗传原因,一组大约 80 种多基因疾病,其特征是健康组织受到攻击的异常免疫反应。尽管序列数据生成已成为常规且负担得起的,但由于目前尚无确定致病变异的金标准方法,仍然存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。目前尚无确定致病变异的金标准方法,因此仍然存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。目前尚无确定致病变异的金标准方法,但仍存在重大挑战。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于为临床实践提供信息。本综述审查了用于识别自身免疫性疾病致病变异的最新方法,并考虑了可用的测序选项和随后的生物信息学方法和策略。开发可靠且稳健的测序和分析工作流程来检测致病变异对于实现精准医学计划的潜力至关重要,在该计划中,患者变异信息用于告知临床实践。


























 京公网安备 11010802027423号
京公网安备 11010802027423号