Journal of Computational Physics ( IF 4.1 ) Pub Date : 2020-07-03 , DOI: 10.1016/j.jcp.2020.109700 Thomas Grandits 1, 2 , Karli Gillette 2, 3 , Aurel Neic 3 , Jason Bayer 4 , Edward Vigmond 4 , Thomas Pock 1, 2 , Gernot Plank 2, 3
|
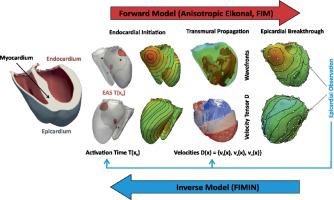
A key mechanism controlling cardiac function is the electrical activation sequence of the heart's main pumping chambers termed the ventricles. As such, personalization of the ventricular activation sequences is of pivotal importance for the clinical utility of computational models of cardiac electrophysiology. However, a direct observation of the activation sequence throughout the ventricular volume is virtually impossible.
In this study, we report on a novel method for identification of activation sequences from activation maps measured at the outer surface of the heart termed the epicardium. Conceptually, the method attempts to identify the key factors governing the ventricular activation sequence – the timing of earliest activation sites (EAS) and the velocity tensor field within the ventricular walls – from sparse and noisy activation maps sampled from the epicardial surface and fits an Eikonal model to the observations.
Regularization methods are first investigated to overcome the severe ill-posedness of the inverse problem in a simplified 2D example. These methods are then employed in an anatomically accurate biventricular model with two realistic activation models of varying complexity – a simplified trifascicular model (3F) and a topologically realistic model of the His-Purkinje system (HPS). Using epicardial activation maps at full resolution, we first demonstrate that reconstructing the volumetric activation sequence is, in principle, feasible under the assumption of known location of EAS and later evaluate robustness of the method against noise and reduced spatial resolution of observations.
Our results suggest that the FIMIN algorithm is able to robustly recover the full 3D activation sequence using epicardial activation maps at a spatial resolution achievable with current mapping systems and in the presence of noise. Comparing the accuracy achieved in the reconstructed activation maps with clinical data uncertainties suggests that the FIMIN method may be suitable for the patient-specific parameterization of activation models.
中文翻译:

从心外膜激活图识别心室激活序列的反电子方法。
控制心脏功能的关键机制是被称为心室的心脏主泵室的电激活顺序。因此,心室激活序列的个性化对于心脏电生理计算模型的临床实用至关重要。但是,实际上不可能直接观察整个心室容积的激活序列。
在这项研究中,我们报告了一种新的方法,该方法可从在称为外膜的心脏外表面测得的激活图识别激活序列。从概念上讲,该方法试图从心外膜表面采样的稀疏和嘈杂的激活图中识别出控制心室激活序列的关键因素-早期激活部位(EAS)的时间和心室壁内的速度张量场-并拟合Eikonal对观察结果建模。
首先研究了正则化方法,以在简化的2D示例中克服反问题的严重不适定性。然后,将这些方法用于解剖学上精确的双心室模型中,该模型具有两个复杂度各不相同的逼真的激活模型-简化的三束模型(3F)和His-Purkinje系统的拓扑逼真的模型(HPS)。使用全分辨率的心外膜激活图,我们首先证明,在已知EAS位置的前提下,重建体积激活序列在原则上是可行的,然后评估该方法抗噪声和降低观测空间分辨率的鲁棒性。
我们的结果表明,FIMIN算法能够使用心外膜激活图以当前成像系统可获得的空间分辨率并在存在噪声的情况下稳健地恢复完整的3D激活序列。将重建的激活图中获得的准确性与临床数据不确定性进行比较,表明FIMIN方法可能适合于特定于患者的激活模型参数化。


























 京公网安备 11010802027423号
京公网安备 11010802027423号