Current Proteomics ( IF 0.8 ) Pub Date : 2020-07-31 , DOI: 10.2174/1570164616666190306151423 Chang Xu 1 , Yijie Ding 2 , Limin Jiang 1 , Cong Shen 1 , Gaoyan Zhang 1 , Xuyao Yu 3
|
Background: The ligand-receptor interaction plays an important role in signal transduction required for cellular differentiation, proliferation, and immune response process. The analysis of ligand-receptor interactions is helpful to provide a deeper understanding of cellular proliferation/ differentiation and other cell processes.
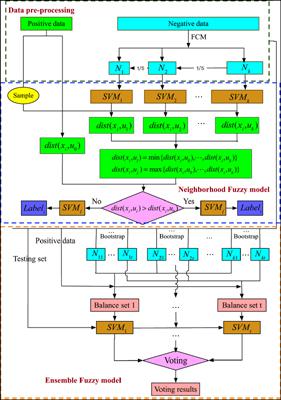
Methods: The computational technique would be used to promote ligand-receptor interactions research in future proteomics research. In this paper, we propose a novel computational method to predict ligand-receptor interactions from amino acid sequences by a machine learning approach. We extract features from ligand and receptor sequences by Histogram of Oriented Gradient (HOG) and Discrete Cosine Transform (DCT). Then, these features are fed into the Fuzzy C-Means (FCM) clustering algorithm for clustering, and also we get multiple training subsets to generate the same number of sub-classifiers. We choose an optimal sub-classifier for predicting ligand-receptor interactions according to the similarity from one sample to training subsets.
Observations: In order to verify the performance, we perform five-fold cross-validation experiments on a ligand-receptor interactions dataset and achieve 80.08% accuracy, 82.98% sensitivity and 80.02% specificity. Then, we test our extracted feature method on two Protein-Protein Interactions (PPIs) datasets, and achieve accuracies of 93.79% and 87.46%, respectively.
Conclusion: Our proposed method can be a useful tool for identifying of ligand-receptor interactions. Related data sets and source code are available at https://github.com/guofei-tju/ligand-receptorinteractions. git.
中文翻译:

通过集成的模糊模型识别配体-受体相互作用
背景:配体-受体相互作用在细胞分化,增殖和免疫反应过程所需的信号转导中起重要作用。配体-受体相互作用的分析有助于深入了解细胞增殖/分化和其他细胞过程。
方法:该计算技术将用于促进未来蛋白质组学研究中的配体-受体相互作用研究。在本文中,我们提出了一种新的计算方法,可以通过机器学习方法来预测氨基酸序列中的配体-受体相互作用。我们通过定向梯度直方图(HOG)和离散余弦变换(DCT)从配体和受体序列中提取特征。然后,将这些特征输入到模糊C均值(FCM)聚类算法中进行聚类,并且我们得到多个训练子集以生成相同数量的子分类器。根据从一个样本到训练子集的相似性,我们选择一个最佳的子分类器来预测配体-受体的相互作用。
观察结果:为了验证性能,我们对配体-受体相互作用数据集进行了五次交叉验证实验,获得了80.08%的准确性,82.98%的敏感性和80.02%的特异性。然后,我们在两个蛋白质-蛋白质相互作用(PPI)数据集上测试提取的特征方法,并分别达到93.79%和87.46%的准确性。
结论:我们提出的方法可以作为鉴定配体-受体相互作用的有用工具。相关数据集和源代码可从https://github.com/guofei-tju/ligand-receptorinteractions获得。git。


























 京公网安备 11010802027423号
京公网安备 11010802027423号