当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Distributed Implementation of Boolean Functions by Transcriptional Synthetic Circuits.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-06-26 , DOI: 10.1021/acssynbio.0c00228 M Ali Al-Radhawi 1 , Anh Phong Tran 2 , Elizabeth A Ernst 3 , Tianchi Chen 4 , Christopher A Voigt 5 , Eduardo D Sontag 1, 4, 6
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-06-26 , DOI: 10.1021/acssynbio.0c00228 M Ali Al-Radhawi 1 , Anh Phong Tran 2 , Elizabeth A Ernst 3 , Tianchi Chen 4 , Christopher A Voigt 5 , Eduardo D Sontag 1, 4, 6
Affiliation
|
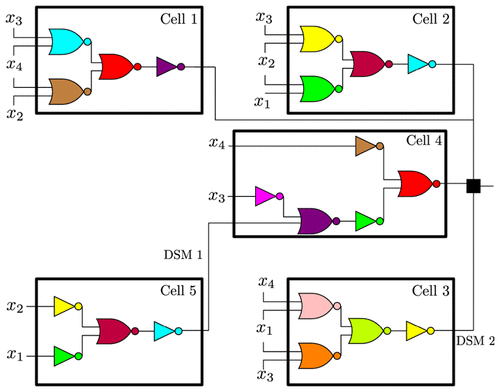
Starting in the early 2000s, sophisticated technologies have been developed for the rational construction of synthetic genetic networks that implement specified logical functionalities. Despite impressive progress, however, the scaling necessary in order to achieve greater computational power has been hampered by many constraints, including repressor toxicity and the lack of large sets of mutually orthogonal repressors. As a consequence, a typical circuit contains no more than roughly seven repressor-based gates per cell. A possible way around this scalability problem is to distribute the computation among multiple cell types, each of which implements a small subcircuit, which communicate among themselves using diffusible small molecules (DSMs). Examples of DSMs are those employed by quorum sensing systems in bacteria. This paper focuses on systematic ways to implement this distributed approach, in the context of the evaluation of arbitrary Boolean functions. The unique characteristics of genetic circuits and the properties of DSMs require the development of new Boolean synthesis methods, distinct from those classically used in electronic circuit design. In this work, we propose a fast algorithm to synthesize distributed realizations for any Boolean function, under constraints on the number of gates per cell and the number of orthogonal DSMs. The method is based on an exact synthesis algorithm to find the minimal circuit per cell, which in turn allows us to build an extensive database of Boolean functions up to a given number of inputs. For concreteness, we will specifically focus on circuits of up to 4 inputs, which might represent, for example, two chemical inducers and two light inputs at different frequencies. Our method shows that, with a constraint of no more than seven gates per cell, the use of a single DSM increases the total number of realizable circuits by at least 7.58-fold compared to centralized computation. Moreover, when allowing two DSM’s, one can realize 99.995% of all possible 4-input Boolean functions, still with at most 7 gates per cell. The methodology introduced here can be readily adapted to complement recent genetic circuit design automation software. A toolbox that uses the proposed algorithm was created and made available at https://github.com/sontaglab/DBC/.
中文翻译:

转录合成电路对布尔函数的分布式实现。
从2000年代初开始,已经开发出复杂的技术来合理构建实现特定逻辑功能的合成遗传网络。尽管取得了令人瞩目的进步,但是为实现更大的计算能力所必需的缩放仍受到许多限制因素的制约,其中包括阻遏物毒性和缺少大量相互正交的阻遏物。结果,一个典型的电路每个单元包含不超过七个基于阻遏器的门。解决此可伸缩性问题的一种可能方法是在多个单元类型之间分配计算,每个单元类型都实现一个小的子电路,这些子电路使用可扩散的小分子(DSM)在它们之间进行通信。DSM的示例是细菌中群体感应系统采用的那些DSM。本文着重于在评估任意布尔函数的背景下实现此分布式方法的系统方法。遗传电路的独特特征和DSM的特性要求开发新的布尔合成方法,这与电子电路设计中传统使用的方法不同。在这项工作中,我们提出了一种快速算法,可以在每个单元的门数和正交DSM数量的约束下,为任何布尔函数综合分布式实现。该方法基于精确的合成算法来找到每个单元的最小电路,这反过来又使我们能够建立一个布尔函数的扩展数据库,直到给定数量的输入。具体而言,我们将特别关注最多4个输入的电路,例如,这可能表示:两个化学诱导剂和两个不同频率的光输入。我们的方法表明,在每个单元不超过七个门的约束下,与集中式计算相比,使用单个DSM可使可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。与集中式计算相比,使用单个DSM可将可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。与集中式计算相比,使用单个DSM可使可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。
更新日期:2020-08-21
中文翻译:

转录合成电路对布尔函数的分布式实现。
从2000年代初开始,已经开发出复杂的技术来合理构建实现特定逻辑功能的合成遗传网络。尽管取得了令人瞩目的进步,但是为实现更大的计算能力所必需的缩放仍受到许多限制因素的制约,其中包括阻遏物毒性和缺少大量相互正交的阻遏物。结果,一个典型的电路每个单元包含不超过七个基于阻遏器的门。解决此可伸缩性问题的一种可能方法是在多个单元类型之间分配计算,每个单元类型都实现一个小的子电路,这些子电路使用可扩散的小分子(DSM)在它们之间进行通信。DSM的示例是细菌中群体感应系统采用的那些DSM。本文着重于在评估任意布尔函数的背景下实现此分布式方法的系统方法。遗传电路的独特特征和DSM的特性要求开发新的布尔合成方法,这与电子电路设计中传统使用的方法不同。在这项工作中,我们提出了一种快速算法,可以在每个单元的门数和正交DSM数量的约束下,为任何布尔函数综合分布式实现。该方法基于精确的合成算法来找到每个单元的最小电路,这反过来又使我们能够建立一个布尔函数的扩展数据库,直到给定数量的输入。具体而言,我们将特别关注最多4个输入的电路,例如,这可能表示:两个化学诱导剂和两个不同频率的光输入。我们的方法表明,在每个单元不超过七个门的约束下,与集中式计算相比,使用单个DSM可使可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。与集中式计算相比,使用单个DSM可将可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。与集中式计算相比,使用单个DSM可使可实现电路的总数增加至少7.58倍。此外,当允许两个DSM时,一个可以实现99.995%的所有可能的4输入布尔函数,每个单元最多仍具有7个门。这里介绍的方法可以很容易地适应最新的遗传电路设计自动化软件。创建了使用建议算法的工具箱,并在https://github.com/sontaglab/DBC/上提供了该工具箱。


























 京公网安备 11010802027423号
京公网安备 11010802027423号