当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Computational Investigation of Structural Interfaces of Protein Complexes with Short Linear Motifs.
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-06-24 , DOI: 10.1021/acs.jproteome.0c00212 Raghavender Surya Upadhyayula 1
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-06-24 , DOI: 10.1021/acs.jproteome.0c00212 Raghavender Surya Upadhyayula 1
Affiliation
|
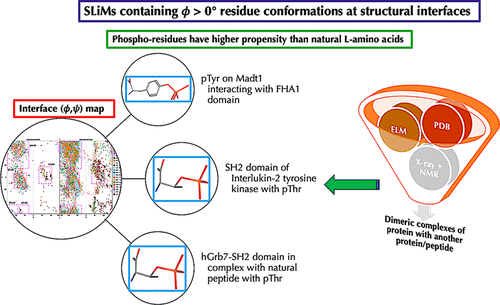
Protein complexes with short linear motifs (SLiMs) are known to play important regulatory functions in eukaryotes. In this investigation, I have studied the structures deposited in PDB with SLiMs. The structures were grouped into three broad categories of protein–protein, protein–peptide, and the rest as others. Protein–peptide complexes were found to be most highly represented. The interfaces were evaluated for geometric features and conformational variables. It was observed that protein–protein and protein–peptide complexes show characteristic differences in residue pairings, which were quantified by evaluating normalized contact residue pairing frequencies. Interface residues adopt characteristic canonical residue conformations in the Ramachandran space, with a pronounced preference for positive ϕ conformations. It was observed that phosphorylated residues have an unusual propensity to adopt the positive ϕ conformations at the interface.
中文翻译:

具有短线性基序的蛋白质复合物的结构界面的计算研究。
已知具有短线性基序(SLiMs)的蛋白质复合物在真核生物中起重要的调节功能。在这项调查中,我研究了使用SLiM沉积在PDB中的结构。这些结构分为三大类蛋白质-蛋白质,蛋白质-肽和其他类别。发现蛋白质-肽复合物的代表性最高。对接口的几何特征和构象变量进行了评估。据观察,蛋白质-蛋白质和蛋白质-肽复合物在残基配对中显示出特征差异,这可以通过评估标准化的接触残基配对频率来量化。界面残基在Ramachandran空间中采用特征性规范残基构象,并特别偏爱正ϕ构象。
更新日期:2020-08-08
中文翻译:

具有短线性基序的蛋白质复合物的结构界面的计算研究。
已知具有短线性基序(SLiMs)的蛋白质复合物在真核生物中起重要的调节功能。在这项调查中,我研究了使用SLiM沉积在PDB中的结构。这些结构分为三大类蛋白质-蛋白质,蛋白质-肽和其他类别。发现蛋白质-肽复合物的代表性最高。对接口的几何特征和构象变量进行了评估。据观察,蛋白质-蛋白质和蛋白质-肽复合物在残基配对中显示出特征差异,这可以通过评估标准化的接触残基配对频率来量化。界面残基在Ramachandran空间中采用特征性规范残基构象,并特别偏爱正ϕ构象。


























 京公网安备 11010802027423号
京公网安备 11010802027423号