当前位置:
X-MOL 学术
›
Microb. Biotechnol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The guanidine thiocyanate‐high EDTA method for total microbial RNA extraction from severely heavy metal‐contaminated soils
Microbial Biotechnology ( IF 5.7 ) Pub Date : 2020-06-23 , DOI: 10.1111/1751-7915.13615 Yaxin Pei 1, 2 , Tursunay Mamtimin 1, 2 , Jing Ji 1, 3 , Aman Khan 1, 3 , Apurva Kakade 1, 2 , Tuoyu Zhou 1, 3 , Zhengsheng Yu 2, 3 , Hajira Zain 1, 3 , Wenzhi Yang 1 , Zhenmin Ling 1, 2, 3 , Wenya Zhang 1 , Yingmei Zhang 1 , Xiangkai Li 1, 2, 3
Microbial Biotechnology ( IF 5.7 ) Pub Date : 2020-06-23 , DOI: 10.1111/1751-7915.13615 Yaxin Pei 1, 2 , Tursunay Mamtimin 1, 2 , Jing Ji 1, 3 , Aman Khan 1, 3 , Apurva Kakade 1, 2 , Tuoyu Zhou 1, 3 , Zhengsheng Yu 2, 3 , Hajira Zain 1, 3 , Wenzhi Yang 1 , Zhenmin Ling 1, 2, 3 , Wenya Zhang 1 , Yingmei Zhang 1 , Xiangkai Li 1, 2, 3
Affiliation
|
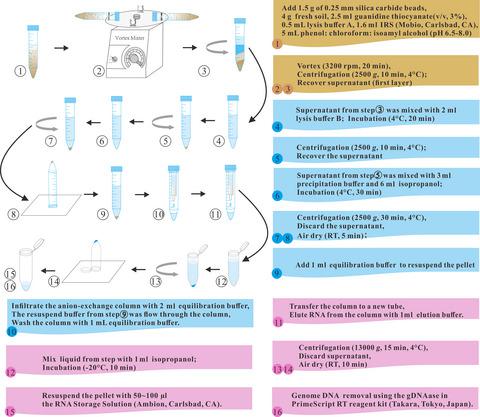
Molecular analyses relying on RNA, as a direct way to unravel active microbes and their functional genes, have received increasing attention from environmental researchers recently. However, extracting sufficient and high‐quality total microbial RNA from seriously heavy metal‐contaminated soils is still a challenge. In this study, the guanidine thiocyanate‐high EDTA (GTHE) method was established and optimized for recovering high quantity and quality of RNA from long‐term heavy metal‐contaminated soils. Due to the low microbial biomass in the soils, we combined multiple strong denaturants and intense mechanical lysis to break cells for increasing RNA yields. To minimize RNAase and heavy metals interference on RNA integrity, the concentrations of guanidine thiocyanate and EDTA were increased from 0.5 to 0.625 ml g−1 soil and 10 to 100 mM, respectively. This optimized GTHE method was applied to seven severely contaminated soils, and the RNA recovery efficiencies were 2.80 ~ 59.41 μg g−1 soil. The total microbial RNA of non‐Cr(VI) (NT) and Cr(VI)‐treated (CT) samples was utilized for molecular analyses. The result of qRT‐PCR demonstrated that the expressions of two tested genes, chrA and yieF, were respectively upregulated 4.12‐ and 62.43‐fold after Cr(VI) treatment. The total microbial RNA extracted from NT and CT samples, respectively, reached to 26.70 μg and 30.75 μg, which were much higher than the required amount (5 μg) for metatranscriptomic library construction. Besides, ratios of mRNA read were more than 86%, which indicated the high‐quality libraries constructed for metatranscriptomic analysis. In summary, the GTHE method is useful to study microbes of contaminated habitats.
中文翻译:

硫氰酸胍-高EDTA法从重金属污染严重的土壤中提取总微生物RNA
最近,依赖RNA的分子分析作为解开活性微生物及其功能基因的直接方法,受到了环境研究人员的越来越多的关注。但是,从重金属污染严重的土壤中提取足够高质量的总微生物RNA仍然是一个挑战。在这项研究中,建立并优化了硫氰酸胍高EDTA(GTHE)方法,以从受重金属污染的长期土壤中回收大量高质量的RNA。由于土壤中微生物的生物量较低,我们结合了多种强变性剂和强烈的机械裂解作用,以破坏细胞,从而提高RNA产量。为了最大程度地减少RNA酶和重金属对RNA完整性的干扰,硫氰酸胍和EDTA的浓度从0.5增加到0.625 ml g -1土壤和10到100 mM。该优化的GTHE方法应用于7种严重污染的土壤,其RNA回收效率为2.80〜59.41μgg -1土壤。非Cr(VI)(NT)和Cr(VI)处理(CT)样品的总微生物RNA用于分子分析。qRT-PCR的结果表明两个测试基因chrA和yieF的表达Cr(VI)处理后,分别上调4.12倍和62.43倍。从NT和CT样品中提取的总微生物RNA分别达到26.70μg和30.75μg,远高于构建超转录组文库所需的量(5μg)。此外,mRNA的读取率超过86%,这表明构建用于元转录组分析的高质量文库。总之,GTHE方法可用于研究受污染生境的微生物。
更新日期:2020-06-23
中文翻译:

硫氰酸胍-高EDTA法从重金属污染严重的土壤中提取总微生物RNA
最近,依赖RNA的分子分析作为解开活性微生物及其功能基因的直接方法,受到了环境研究人员的越来越多的关注。但是,从重金属污染严重的土壤中提取足够高质量的总微生物RNA仍然是一个挑战。在这项研究中,建立并优化了硫氰酸胍高EDTA(GTHE)方法,以从受重金属污染的长期土壤中回收大量高质量的RNA。由于土壤中微生物的生物量较低,我们结合了多种强变性剂和强烈的机械裂解作用,以破坏细胞,从而提高RNA产量。为了最大程度地减少RNA酶和重金属对RNA完整性的干扰,硫氰酸胍和EDTA的浓度从0.5增加到0.625 ml g -1土壤和10到100 mM。该优化的GTHE方法应用于7种严重污染的土壤,其RNA回收效率为2.80〜59.41μgg -1土壤。非Cr(VI)(NT)和Cr(VI)处理(CT)样品的总微生物RNA用于分子分析。qRT-PCR的结果表明两个测试基因chrA和yieF的表达Cr(VI)处理后,分别上调4.12倍和62.43倍。从NT和CT样品中提取的总微生物RNA分别达到26.70μg和30.75μg,远高于构建超转录组文库所需的量(5μg)。此外,mRNA的读取率超过86%,这表明构建用于元转录组分析的高质量文库。总之,GTHE方法可用于研究受污染生境的微生物。



























 京公网安备 11010802027423号
京公网安备 11010802027423号