当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Parameterization of metabolite and macromolecule contributions in interrelated MR spectra of human brain using multidimensional modeling.
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-06-15 , DOI: 10.1002/nbm.4328 Maike Hoefemann 1, 2 , Christine Sandra Bolliger 1, 3 , Daniel G Q Chong 1, 2 , Jan Willem van der Veen 4 , Roland Kreis 1
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-06-15 , DOI: 10.1002/nbm.4328 Maike Hoefemann 1, 2 , Christine Sandra Bolliger 1, 3 , Daniel G Q Chong 1, 2 , Jan Willem van der Veen 4 , Roland Kreis 1
Affiliation
|
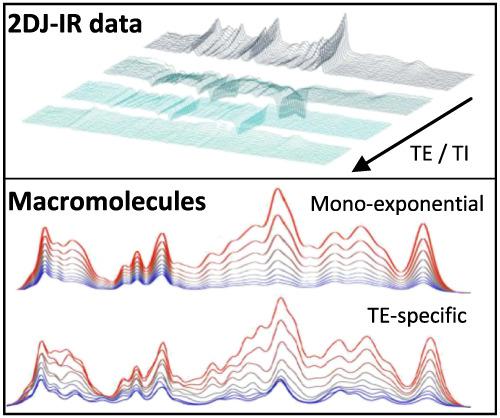
Macromolecular signals are crucial constituents of short echo‐time 1H MR spectra with potential clinical implications in themselves as well as essential ramifications for the quantification of the usually targeted metabolites. Their parameterization, needed for general fitting models, is difficult because of their unknown composition. Here, a macromolecular signal parameterization together with metabolite signal quantification including relaxation properties is investigated by multidimensional modeling of interrelated 2DJ inversion‐recovery (2DJ‐IR) datasets. Simultaneous and iterative procedures for defining the macromolecular background (MMBG) as mono‐exponentially or generally decaying signals over TE are evaluated. Varying prior knowledge and restrictions in the metabolite evaluation are tested to examine their impact on results and fitting stability for two sets of three‐dimensional spectra acquired with metabolite‐cycled PRESS from cerebral gray and white matter locations. One dataset was used for model optimization, and also examining the influence of prior knowledge on estimated parameters. The most promising model was applied to a second dataset. It turned out that the mono‐exponential decay model appears to be inadequate to represent TE‐dependent signal features of the MMBG. TE‐adapted MMBG spectra were therefore determined. For a reliable overall quantification of implicated metabolite concentrations and relaxation times, a general fitting model had to be constrained in terms of the number of fitting variables and the allowed parameter space. With such a model in place, fitting precision for metabolite contents and relaxation times was excellent, while fitting accuracy is difficult to judge and bias was likely influenced by the type of fitting constraints enforced. In summary, the parameterization of metabolite and macromolecule contributions in interrelated MR spectra has been examined by using multidimensional modeling on complex 2DJ‐IR datasets. A tightly restricted model allows fitting of individual subject data with high fitting precision documented in small Cramér‐Rao lower bounds, good repeatability values and a relatively small spread of estimated concentration and relaxation values for a healthy subject cohort.
中文翻译:

使用多维建模对人脑相关 MR 谱中的代谢物和大分子贡献进行参数化。
大分子信号是短回波时间的关键组成部分1H MR 谱本身具有潜在的临床意义,以及对通常目标代谢物的定量分析的基本结果。由于它们的组成未知,因此一般拟合模型所需的参数化很困难。在这里,通过相互关联的 2DJ 反转恢复 (2DJ-IR) 数据集的多维建模研究了大分子信号参数化以及包括弛豫特性在内的代谢物信号量化。评估了用于将大分子背景 (MMBG) 定义为 TE 上的单指数或一般衰减信号的同时和迭代程序。对代谢物评估中的不同先验知识和限制进行了测试,以检查它们对使用代谢物循环 PRESS 从大脑灰质和白质位置获得的两组三维光谱的结果和拟合稳定性的影响。一个数据集用于模型优化,并检查先验知识对估计参数的影响。最有前途的模型被应用于第二个数据集。事实证明,单指数衰减模型似乎不足以表示 MMBG 的 TE 相关信号特征。因此确定了 TE 适应的 MMBG 光谱。为了对相关代谢物浓度和弛豫时间进行可靠的整体量化,必须在拟合变量的数量和允许的参数空间方面限制通用拟合模型。有了这样的模型,代谢物含量和弛豫时间的拟合精度非常好,而拟合精度很难判断,偏差可能会受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。而拟合的准确性很难判断,而且偏差很可能受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。而拟合的准确性很难判断,而且偏差很可能受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。
更新日期:2020-08-04
中文翻译:

使用多维建模对人脑相关 MR 谱中的代谢物和大分子贡献进行参数化。
大分子信号是短回波时间的关键组成部分1H MR 谱本身具有潜在的临床意义,以及对通常目标代谢物的定量分析的基本结果。由于它们的组成未知,因此一般拟合模型所需的参数化很困难。在这里,通过相互关联的 2DJ 反转恢复 (2DJ-IR) 数据集的多维建模研究了大分子信号参数化以及包括弛豫特性在内的代谢物信号量化。评估了用于将大分子背景 (MMBG) 定义为 TE 上的单指数或一般衰减信号的同时和迭代程序。对代谢物评估中的不同先验知识和限制进行了测试,以检查它们对使用代谢物循环 PRESS 从大脑灰质和白质位置获得的两组三维光谱的结果和拟合稳定性的影响。一个数据集用于模型优化,并检查先验知识对估计参数的影响。最有前途的模型被应用于第二个数据集。事实证明,单指数衰减模型似乎不足以表示 MMBG 的 TE 相关信号特征。因此确定了 TE 适应的 MMBG 光谱。为了对相关代谢物浓度和弛豫时间进行可靠的整体量化,必须在拟合变量的数量和允许的参数空间方面限制通用拟合模型。有了这样的模型,代谢物含量和弛豫时间的拟合精度非常好,而拟合精度很难判断,偏差可能会受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。而拟合的准确性很难判断,而且偏差很可能受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。而拟合的准确性很难判断,而且偏差很可能受到强制执行的拟合约束类型的影响。总之,通过在复杂的 2DJ-IR 数据集上使用多维建模,已经检查了相关 MR 光谱中代谢物和大分子贡献的参数化。严格限制的模型允许以小 Cramér-Rao 下限、良好的可重复性值和健康受试者队列的估计浓度和松弛值的相对较小的范围记录的高拟合精度来拟合个体受试者数据。



























 京公网安备 11010802027423号
京公网安备 11010802027423号