Current Bioinformatics ( IF 4 ) Pub Date : 2020-05-01 , DOI: 10.2174/1574893614666191018171521 Wei Chen 1 , Kewei Liu 1
|
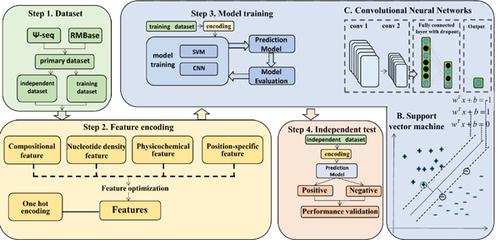
Background: Pseudouridine (Ψ) is the most abundant RNA modification and has important functions in a series of biological and cellular processes. Although experimental techniques have made great contributions to identify Ψ sites, they are still labor-intensive and costineffective. In the past few years, a series of computational approaches have been developed, which provided rapid and efficient approaches to identify Ψ sites.
Results: To provide the readership with a clear landscape about the recent development in this important area, in this review, we summarized and compared the representative computational approaches developed for identifying Ψ sites. Moreover, future directions in computationally identifying Ψ sites were discussed as well.
Conclusion: We anticipate that this review will provide novel insights into the researches on pseudouridine modification.
中文翻译:

RNA假尿苷位点预测工具的分析和比较
背景:假性尿苷(Ψ)是最丰富的RNA修饰,在一系列生物学和细胞过程中具有重要功能。尽管实验技术为识别Ψ点做出了巨大贡献,但它们仍然是劳动密集型且成本低廉的。在过去的几年中,已经开发出了一系列计算方法,这些方法提供了快速有效的方法来识别Ψ点。
结果:为了向读者提供有关该重要领域最近发展的清晰情况,在本综述中,我们总结并比较了为识别Ψ站点而开发的代表性计算方法。此外,还讨论了在计算上识别Ψ位点的未来方向。
结论:我们预计该综述将为假尿苷修饰的研究提供新颖的见解。


























 京公网安备 11010802027423号
京公网安备 11010802027423号