当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Genomic signals found using RNA sequencing show signatures of selection and subtle population differentiation in walleye (Sander vitreus) in a large freshwater ecosystem.
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-06-13 , DOI: 10.1002/ece3.6418 Matt J Thorstensen 1 , Jennifer D Jeffrey 1 , Jason R Treberg 1 , Douglas A Watkinson 2 , Eva C Enders 2 , Ken M Jeffries 1
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-06-13 , DOI: 10.1002/ece3.6418 Matt J Thorstensen 1 , Jennifer D Jeffrey 1 , Jason R Treberg 1 , Douglas A Watkinson 2 , Eva C Enders 2 , Ken M Jeffries 1
Affiliation
|
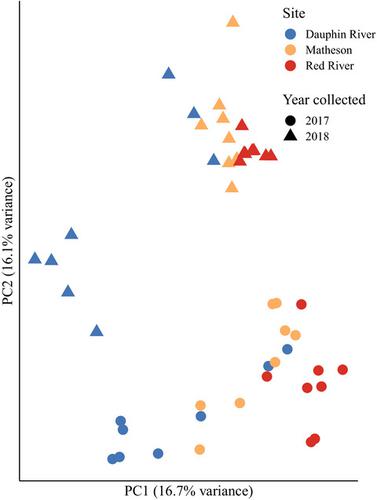
RNA sequencing is an effective approach for studying aquatic species yielding both physiological and genomic data. However, its population genetic applications are not well‐characterized. We investigate this possible role for RNA sequencing for population genomics in Lake Winnipeg, Manitoba, Canada, walleye (Sander vitreus ). Lake Winnipeg walleye represent the largest component of the second‐largest freshwater fishery in Canada. In the present study, large female walleye were sampled via nonlethal gill biopsy over two years at three spawning sites representing a latitudinal gradient in the lake. Genetic variation from sequenced mRNA was analyzed for neutral and adaptive markers to investigate population structure and possible adaptive variation. We find low population divergence (F ST = 0.0095), possible northward gene flow, and outlier loci that vary latitudinally in transcripts associated with cell membrane proteins and cytoskeletal function. These results indicate that Lake Winnipeg walleye may be effectively managed as a single demographically connected metapopulation with contributing subpopulations and suggest genomic differences possibly underlying observed phenotypic differences. Despite its high cost relative to other genotyping methods, RNA sequencing data can yield physiological in addition to genetic information discussed here. We therefore argue that it is useful for addressing diverse molecular questions in the conservation of freshwater species.
中文翻译:

使用RNA测序发现的基因组信号显示出在大型淡水生态系统中的角膜白斑(沙丘玻璃体)中的选择和微妙的种群分化特征。
RNA测序是研究产生生理和基因组数据的水生物种的有效方法。但是,它的种群遗传应用没有很好的特征。我们调查了温尼伯湖,曼尼托巴省,加拿大,角膜白斑(Sander vitreus)的种群基因组学RNA测序的这一可能作用。温尼伯湖角膜白斑鱼是加拿大第二大淡水渔业的最大组成部分。在本研究中,通过非致死性ill活检在两年内在代表湖中纬度梯度的三个产卵地点取样了大型雌性角膜白斑。分析了测序mRNA的遗传变异以寻找中性和适应性标记,以研究种群结构和可能的适应性变异。我们发现人口差异较低(FST = 0.0095),可能的北向基因流以及离群基因座,这些离群基因在与细胞膜蛋白和细胞骨架功能相关的转录本中呈横向变化。这些结果表明,温尼伯湖角膜白斑病可以有效地处理为具有相关亚群的人口统计学联系的亚群,并暗示可能是潜在表型差异的基因组差异。尽管相对于其他基因分型方法而言,其成本很高,但RNA测序数据除了此处讨论的遗传信息外,还可以产生生理学信息。因此,我们认为这对于解决淡水物种保护中的各种分子问题很有用。
更新日期:2020-07-30
中文翻译:

使用RNA测序发现的基因组信号显示出在大型淡水生态系统中的角膜白斑(沙丘玻璃体)中的选择和微妙的种群分化特征。
RNA测序是研究产生生理和基因组数据的水生物种的有效方法。但是,它的种群遗传应用没有很好的特征。我们调查了温尼伯湖,曼尼托巴省,加拿大,角膜白斑(Sander vitreus)的种群基因组学RNA测序的这一可能作用。温尼伯湖角膜白斑鱼是加拿大第二大淡水渔业的最大组成部分。在本研究中,通过非致死性ill活检在两年内在代表湖中纬度梯度的三个产卵地点取样了大型雌性角膜白斑。分析了测序mRNA的遗传变异以寻找中性和适应性标记,以研究种群结构和可能的适应性变异。我们发现人口差异较低(FST = 0.0095),可能的北向基因流以及离群基因座,这些离群基因在与细胞膜蛋白和细胞骨架功能相关的转录本中呈横向变化。这些结果表明,温尼伯湖角膜白斑病可以有效地处理为具有相关亚群的人口统计学联系的亚群,并暗示可能是潜在表型差异的基因组差异。尽管相对于其他基因分型方法而言,其成本很高,但RNA测序数据除了此处讨论的遗传信息外,还可以产生生理学信息。因此,我们认为这对于解决淡水物种保护中的各种分子问题很有用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号