Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2020-05-29 , DOI: 10.1016/j.csbj.2020.05.023 Joon-Yong Lee 1 , Natalie C Sadler 1 , Robert G Egbert 1 , Christopher R Anderton 2 , Kirsten S Hofmockel 2, 3 , Janet K Jansson 1 , Hyun-Seob Song 1, 4, 5
|
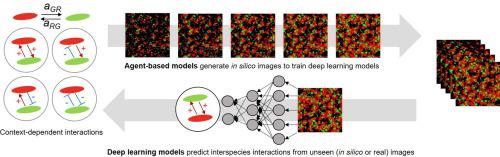
Microbial communities organize into spatial patterns that are largely governed by interspecies interactions. This phenomenon is an important metric for understanding community functional dynamics, yet the use of spatial patterns for predicting microbial interactions is currently lacking. Here we propose supervised deep learning as a new tool for network inference. An agent-based model was used to simulate the spatiotemporal evolution of two interacting organisms under diverse growth and interaction scenarios, the data of which was subsequently used to train deep neural networks. For small-size domains (100 µm × 100 µm) over which interaction coefficients are assumed to be invariant, we obtained fairly accurate predictions, as indicated by an average R2 value of 0.84. In application to relatively larger domains (450 µm × 450 µm) where interaction coefficients are varying in space, deep learning models correctly predicted spatial distributions of interaction coefficients without any additional training. Lastly, we evaluated our model against real biological data obtained using Pseudomonas fluorescens and Escherichia coli co-cultures treated with polymeric chitin or N-acetylglucosamine, the hydrolysis product of chitin. While P. fluorescens can utilize both substrates for growth, E. coli lacked the ability to degrade chitin. Consistent with our expectations, our model predicted context-dependent interactions across two substrates, i.e., degrader-cheater relationship on chitin polymers and competition on monomers. The combined use of the agent-based model and machine learning algorithm successfully demonstrates how to infer microbial interactions from spatially distributed data, presenting itself as a useful tool for the analysis of more complex microbial community interactions.
中文翻译:

深度学习根据自组织的时空模式预测微生物的相互作用。
微生物群落组织成主要由种间相互作用控制的空间格局。这种现象是了解社区功能动力学的重要指标,但是目前缺乏使用空间模式预测微生物相互作用的方法。在这里,我们建议将监督深度学习作为网络推理的新工具。基于代理的模型用于模拟两种相互作用生物在不同生长和相互作用情况下的时空演化,其数据随后用于训练深度神经网络。对于假定相互作用系数不变的小尺寸域(100 µm×100 µm),我们得到了相当准确的预测,如平均R 2所示值为0.84。在交互系数在空间中变化的相对较大的域(450 µm×450 µm)中,深度学习模型可以正确预测交互系数的空间分布,而无需任何额外的训练。最后,我们针对使用荧光假单胞菌和聚合几丁质或几丁质水解产物N-乙酰氨基葡萄糖处理的大肠杆菌共培养获得的真实生物学数据评估了我们的模型。虽然荧光假单胞菌可以利用生长两个基板,大肠杆菌缺乏降解几丁质的能力。与我们的预期一致,我们的模型预测了两种底物之间的上下文相关相互作用,即几丁质聚合物上的降解物-骗子关系和单体上的竞争。基于代理的模型和机器学习算法的结合使用成功地演示了如何从空间分布的数据推断微生物的相互作用,并将其作为分析更复杂的微生物群落相互作用的有用工具。

























 京公网安备 11010802027423号
京公网安备 11010802027423号