当前位置:
X-MOL 学术
›
Mol. Pharmaceutics
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PTML Model for Selection of Nanoparticles, Anticancer Drugs, and Vitamins in the Design of Drug-Vitamin Nanoparticle Release Systems for Cancer Cotherapy.
Molecular Pharmaceutics ( IF 4.9 ) Pub Date : 2020-05-27 , DOI: 10.1021/acs.molpharmaceut.0c00308 Ricardo Santana 1, 2, 3 , Robin Zuluaga 4 , Piedad Gañán 3 , Sonia Arrasate 5 , Enrique Onieva 2 , Matthew M Montemore 1 , Humbert González-Díaz 5, 6, 7
Molecular Pharmaceutics ( IF 4.9 ) Pub Date : 2020-05-27 , DOI: 10.1021/acs.molpharmaceut.0c00308 Ricardo Santana 1, 2, 3 , Robin Zuluaga 4 , Piedad Gañán 3 , Sonia Arrasate 5 , Enrique Onieva 2 , Matthew M Montemore 1 , Humbert González-Díaz 5, 6, 7
Affiliation
|
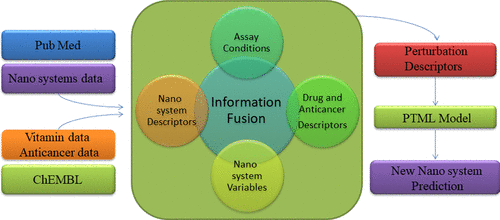
Nanosystems are gaining momentum in pharmaceutical sciences because of the wide variety of possibilities for designing these systems to have specific functions. Specifically, studies of new cancer cotherapy drug–vitamin release nanosystems (DVRNs) including anticancer compounds and vitamins or vitamin derivatives have revealed encouraging results. However, the number of possible combinations of design and synthesis conditions is remarkably high. In addition, a large number of anticancer and vitamin derivatives have been already assayed, but a notably less number of cases of DVRNs were assayed as a whole (with the anticancer compound and the vitamin linked to them). Our approach combines with the perturbation theory and machine learning (PTML) model to predict the probability of obtaining an interesting DVRN by changing the anticancer compound and/or the vitamin present in a DVRN that is already tested for other anticancer compounds or vitamins that have not been tested yet as part of a DVRN. In a previous work, we built a linear PTML model useful for the design of these nanosystems. In doing so, we used information fusion (IF) techniques to carry out data enrichment of DVRN data compiled from the literature with the data for preclinical assays of vitamins from the ChEMBL database. The design features of DVRNs and the assay conditions of nanoparticles (NPs) and vitamins were included as multiplicative PT operators (PTOs) to the system, which indicates the importance of these variables. However, the previous work omitted experiments with nonlinear ML techniques and different types of PTOs such as metric-based PTOs. More importantly, the previous work does not consider the structure of the anticancer drug to be included in the new DVRNs. In this work, we are going to accomplish three main objectives (tasks). In the first task, we found a new model, alternative to the one published before, for the rational design of DVRNs using metric-based PTOs. The most accurate PTML model was the artificial neural network model, which showed values of specificity, sensitivity, and accuracy in the range of 90–95% in training and external validation series for more than 130,000 cases (DVRNs vs ChEMBL assays). Furthermore, in the second task, we used IF techniques to carry out data enrichment of our previous data set. In doing so, we constructed a new working data set of >970,000 cases with the data of preclinical assays of DVRNs, vitamins, and anticancer compounds from the ChEMBL database. All these assays have multiple continuous variables or descriptors dk and categorical variables cj (conditions of the assays) for drugs (dack, cacj), vitamins (dvk, cvj), and NPs (dnk, cnj). These data include >20,000 potential anticancer compounds with >270 protein targets (cac1), >580 assay cell organisms (cac2), and so forth. Furthermore, we include >36,000 assay vitamin derivatives in >6200 types of cells (c2vit), >120 assay organisms (c3vit), >60 assay strains (c4vit), and so forth. The enriched data set also contains >20 types of DVRNs (c5n) with 9 NP core materials (c4n), 8 synthesis methods (c7n), and so forth. We expressed all this information with PTOs and developed a qualitatively new PTML model that incorporates information of the anticancer drugs. This new model presents 96–97% of accuracy for training and external validation subsets. In the last task, we carried out a comparative study of ML and/or PTML models published and described how the models we are presenting cover the gap of knowledge in terms of drug delivery. In conclusion, we present here for the first time a multipurpose PTML model that is able to select NPs, anticancer compounds, and vitamins and their conditions of assay for DVRN design.
中文翻译:

用于癌症共疗法的药物-维生素纳米颗粒释放系统设计中用于选择纳米颗粒,抗癌药物和维生素的PTML模型。
由于将这些系统设计为具有特定功能的可能性多种多样,因此纳米系统在制药科学中正获得发展动力。具体而言,对包括抗癌化合物和维生素或维生素衍生物在内的新型癌症辅助疗法药物维生素释放纳米系统(DVRN)的研究显示出令人鼓舞的结果。然而,设计和合成条件的可能组合的数量非常多。此外,已经对大量的抗癌和维生素衍生物进行了测定,但是从整体上分析了数量较少的DVRNs(将抗癌化合物和维生素与它们相连)。我们的方法与摄动理论和机器学习(PTML)模型相结合,通过更改已针对其他抗癌化合物或未经测试的抗癌化合物或维生素进行测试的DVRN中存在的抗癌化合物和/或维生素来预测获得有趣的DVRN的可能性已作为DVRN的一部分进行了测试。在先前的工作中,我们建立了线性PTML模型,可用于设计这些纳米系统。在此过程中,我们使用信息融合(IF)技术对从文献中汇编的DVRN数据与来自ChEMBL数据库的维生素进行临床前测定的数据进行了数据丰富。DVRNs的设计特征以及纳米颗粒(NPs)和维生素的测定条件作为系统的乘法PT运算符(PTO)包括在内,这表明了这些变量的重要性。然而,先前的工作省略了使用非线性ML技术和不同类型的PTO(例如基于度量的PTO)进行的实验。更重要的是,先前的工作并未考虑将抗癌药物的结构包括在新的DVRN中。在这项工作中,我们将完成三个主要目标(任务)。在第一个任务中,我们找到了一种新模型,用于替代以前发布的模型,该模型用于使用基于度量的PTO合理设计DVRN。最准确的PTML模型是人工神经网络模型,该模型在超过130,000例病例(DVRN与ChEMBL分析)的训练和外部验证系列中显示的特异性,敏感性和准确性值在90-95%的范围内。此外,在第二项任务中,我们使用了IF技术对以前的数据集进行数据丰富。在这样做,我们使用ChEMBL数据库中的DVRNs,维生素和抗癌化合物的临床前测定数据构建了一个大于970,000例的新工作数据集。所有这些测定都有多个连续变量或描述符d k和用于药物(d ac k,c ac j),维生素(d v k,c v j)和NPs(d n k,c n j)的分类变量c j(测定条件)。这些数据包括具有> 270个蛋白靶标(c ac1),> 580个分析细胞生物(c ac2)等> 20,000种潜在的抗癌化合物。此外,我们在> 6200种类型的细胞(c 2vit)中包括了> 36,000种维生素衍生物),> 120种检测生物(c 3vit),> 60种检测菌株(c 4vit)等。丰富的数据集还包含20种以上的DVRN(c 5n)和9种NP核心材料(c 4n),8种合成方法(c 7n),依此类推。我们用PTO表达了所有这些信息,并开发了定性性新的PTML模型,其中包含了抗癌药物的信息。这个新模型提供了训练和外部验证子集的准确度的96–97%。在最后一个任务中,我们对发布的ML和/或PTML模型进行了比较研究,并描述了我们所提供的模型如何覆盖药物输送方面的知识空白。总而言之,我们在这里首次展示了一种多功能PTML模型,该模型能够选择NP,抗癌化合物和维生素及其用于DVRN设计的分析条件。
更新日期:2020-07-06
中文翻译:

用于癌症共疗法的药物-维生素纳米颗粒释放系统设计中用于选择纳米颗粒,抗癌药物和维生素的PTML模型。
由于将这些系统设计为具有特定功能的可能性多种多样,因此纳米系统在制药科学中正获得发展动力。具体而言,对包括抗癌化合物和维生素或维生素衍生物在内的新型癌症辅助疗法药物维生素释放纳米系统(DVRN)的研究显示出令人鼓舞的结果。然而,设计和合成条件的可能组合的数量非常多。此外,已经对大量的抗癌和维生素衍生物进行了测定,但是从整体上分析了数量较少的DVRNs(将抗癌化合物和维生素与它们相连)。我们的方法与摄动理论和机器学习(PTML)模型相结合,通过更改已针对其他抗癌化合物或未经测试的抗癌化合物或维生素进行测试的DVRN中存在的抗癌化合物和/或维生素来预测获得有趣的DVRN的可能性已作为DVRN的一部分进行了测试。在先前的工作中,我们建立了线性PTML模型,可用于设计这些纳米系统。在此过程中,我们使用信息融合(IF)技术对从文献中汇编的DVRN数据与来自ChEMBL数据库的维生素进行临床前测定的数据进行了数据丰富。DVRNs的设计特征以及纳米颗粒(NPs)和维生素的测定条件作为系统的乘法PT运算符(PTO)包括在内,这表明了这些变量的重要性。然而,先前的工作省略了使用非线性ML技术和不同类型的PTO(例如基于度量的PTO)进行的实验。更重要的是,先前的工作并未考虑将抗癌药物的结构包括在新的DVRN中。在这项工作中,我们将完成三个主要目标(任务)。在第一个任务中,我们找到了一种新模型,用于替代以前发布的模型,该模型用于使用基于度量的PTO合理设计DVRN。最准确的PTML模型是人工神经网络模型,该模型在超过130,000例病例(DVRN与ChEMBL分析)的训练和外部验证系列中显示的特异性,敏感性和准确性值在90-95%的范围内。此外,在第二项任务中,我们使用了IF技术对以前的数据集进行数据丰富。在这样做,我们使用ChEMBL数据库中的DVRNs,维生素和抗癌化合物的临床前测定数据构建了一个大于970,000例的新工作数据集。所有这些测定都有多个连续变量或描述符d k和用于药物(d ac k,c ac j),维生素(d v k,c v j)和NPs(d n k,c n j)的分类变量c j(测定条件)。这些数据包括具有> 270个蛋白靶标(c ac1),> 580个分析细胞生物(c ac2)等> 20,000种潜在的抗癌化合物。此外,我们在> 6200种类型的细胞(c 2vit)中包括了> 36,000种维生素衍生物),> 120种检测生物(c 3vit),> 60种检测菌株(c 4vit)等。丰富的数据集还包含20种以上的DVRN(c 5n)和9种NP核心材料(c 4n),8种合成方法(c 7n),依此类推。我们用PTO表达了所有这些信息,并开发了定性性新的PTML模型,其中包含了抗癌药物的信息。这个新模型提供了训练和外部验证子集的准确度的96–97%。在最后一个任务中,我们对发布的ML和/或PTML模型进行了比较研究,并描述了我们所提供的模型如何覆盖药物输送方面的知识空白。总而言之,我们在这里首次展示了一种多功能PTML模型,该模型能够选择NP,抗癌化合物和维生素及其用于DVRN设计的分析条件。



























 京公网安备 11010802027423号
京公网安备 11010802027423号