Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Phylogenomic analysis of the APETALA2 transcription factor subfamily across angiosperms reveals both deep conservation and lineage-specific patterns.
The Plant Journal ( IF 7.2 ) Pub Date : 2020-05-20 , DOI: 10.1111/tpj.14843 Merijn H L Kerstens 1 , M Eric Schranz 1 , Klaas Bouwmeester 1
The Plant Journal ( IF 7.2 ) Pub Date : 2020-05-20 , DOI: 10.1111/tpj.14843 Merijn H L Kerstens 1 , M Eric Schranz 1 , Klaas Bouwmeester 1
Affiliation
|
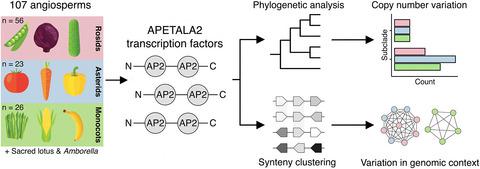
The APETALA2 (AP2) subfamily of transcription factors are key regulators of angiosperm root, shoot, flower and embryo development. The broad diversity of anatomical and morphological structures is potentially associated with the genomic dynamics of the AP2 subfamily. However, a comprehensive phylogenomic analysis of the AP2 subfamily across angiosperms is lacking. We combined phylogenetic and synteny analysis of distinct AP2 subclades in the completed genomes of 107 angiosperm species. We identified major changes in copy number variation and genomic context within subclades across lineages, and discuss how these changes may have contributed to the evolution of lineage‐specific traits. Multiple AP2 subclades show highly conserved patterns of copy number and synteny across angiosperms, while others are more dynamic and show distinct lineage‐specific patterns. As examples of lineage‐specific morphological divergence due to AP2 subclade dynamics, we hypothesize that loss of PLETHORA1/2 in monocots correlates with the absence of taproots, whereas independent lineage‐specific changes of PLETHORA4/BABY BOOM and WRINKLED1 genes in Brassicaceae and monocots point towards regulatory divergence of embryogenesis between these lineages. Additionally, copy number expansion of TOE1 and TOE3/AP2 in asterids is implicated with differential regulation of flower development. Moreover, we show that the genomic context of AP2s is in general highly specialized per angiosperm lineage. To our knowledge, this study is the first to shed light on the evolutionary divergence of the AP2 subfamily subclades across major angiosperm lineages and emphasizes the need for lineage‐specific characterization of developmental networks to understand trait variability further.
中文翻译:

跨被子植物的APETALA2转录因子亚家族的系统生物学分析揭示了深度保守和谱系特异性模式。
转录因子的APETALA2(AP2)亚家族是被子植物根,茎,花和胚胎发育的关键调控因子。解剖结构和形态结构的广泛多样性可能与AP2亚家族的基因组动力学有关。但是,缺乏对整个被子植物AP2亚科的全面系统生物学分析。我们结合了107个被子植物完整基因组中不同AP2子代的系统发育和同义分析。我们确定了跨谱系亚群内拷贝数变异和基因组背景的主要变化,并讨论了这些变化如何可能有助于谱系特质的进化。多个AP2子瓣在被子植物中显示出高度保守的拷贝数和同构模式,而其他子囊则更具动态性,并表现出独特的谱系特异性模式。作为AP2子代动力学导致的特定谱系形态差异的例子,我们假设单子叶植物中PLETHORA1 / 2的丧失与主根的缺失相关,而十字花科和单子叶植物中PLETHORA4 / BABY BOOM和WRINKLED1基因的独立于谱系特定的变化这些谱系之间的胚胎发生调控差异。此外,TOE1和TOE3 / AP2的副本号扩展在星状星体中,与花发育的差异调节有关。此外,我们表明,AP2 s的基因组背景通常是每个被子植物谱系高度专门化的。据我们所知,本研究首次揭示了主要被子植物谱系中AP2亚科亚群在进化上的差异,并强调需要对谱系进行特定的表征,以进一步了解性状变异性。
更新日期:2020-05-20
中文翻译:

跨被子植物的APETALA2转录因子亚家族的系统生物学分析揭示了深度保守和谱系特异性模式。
转录因子的APETALA2(AP2)亚家族是被子植物根,茎,花和胚胎发育的关键调控因子。解剖结构和形态结构的广泛多样性可能与AP2亚家族的基因组动力学有关。但是,缺乏对整个被子植物AP2亚科的全面系统生物学分析。我们结合了107个被子植物完整基因组中不同AP2子代的系统发育和同义分析。我们确定了跨谱系亚群内拷贝数变异和基因组背景的主要变化,并讨论了这些变化如何可能有助于谱系特质的进化。多个AP2子瓣在被子植物中显示出高度保守的拷贝数和同构模式,而其他子囊则更具动态性,并表现出独特的谱系特异性模式。作为AP2子代动力学导致的特定谱系形态差异的例子,我们假设单子叶植物中PLETHORA1 / 2的丧失与主根的缺失相关,而十字花科和单子叶植物中PLETHORA4 / BABY BOOM和WRINKLED1基因的独立于谱系特定的变化这些谱系之间的胚胎发生调控差异。此外,TOE1和TOE3 / AP2的副本号扩展在星状星体中,与花发育的差异调节有关。此外,我们表明,AP2 s的基因组背景通常是每个被子植物谱系高度专门化的。据我们所知,本研究首次揭示了主要被子植物谱系中AP2亚科亚群在进化上的差异,并强调需要对谱系进行特定的表征,以进一步了解性状变异性。


























 京公网安备 11010802027423号
京公网安备 11010802027423号