当前位置:
X-MOL 学术
›
Mol. Plant Pathol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Development of FRET-based high-throughput screening for viral RNase III inhibitors.
Molecular Plant Pathology ( IF 4.9 ) Pub Date : 2020-05-21 , DOI: 10.1111/mpp.12942 Linping Wang 1 , Jani Saarela 2 , Sylvain Poque 1 , Jari P T Valkonen 1
Molecular Plant Pathology ( IF 4.9 ) Pub Date : 2020-05-21 , DOI: 10.1111/mpp.12942 Linping Wang 1 , Jani Saarela 2 , Sylvain Poque 1 , Jari P T Valkonen 1
Affiliation
|
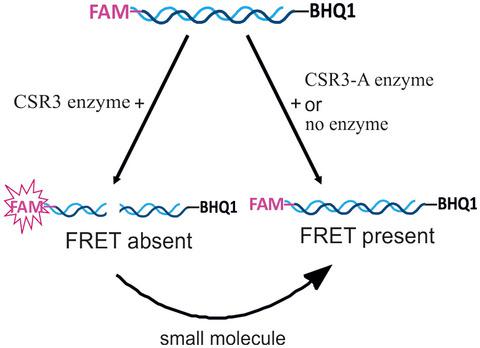
The class 1 ribonuclease III (RNase III) encoded by Sweet potato chlorotic stunt virus (CSR3) suppresses RNA silencing in plant cells and thereby counters the host antiviral response by cleaving host small interfering RNAs, which are indispensable components of the plant RNA interference (RNAi) pathway. The synergy between sweet potato chlorotic stunt virus and sweet potato feathery mottle virus can reduce crop yields by 90%. Inhibitors of CSR3 might prove efficacious to counter this viral threat, yet no screen has been carried out to identify such inhibitors. Here, we report a novel high‐throughput screening (HTS) assay based on fluorescence resonance energy transfer (FRET) for identifying inhibitors of CSR3. For monitoring CSR3 activity via HTS, we used a small interfering RNA substrate that was labelled with a FRET‐compatible dye. The optimized HTS assay yielded 109 potential inhibitors of CSR3 out of 6,620 compounds tested from different small‐molecule libraries. The three best inhibitor candidates were validated with a dose–response assay. In addition, a parallel screen of the selected candidates was carried out for a similar class 1 RNase III enzyme from Escherichia coli (EcR3), and this screen yielded a different set of inhibitors. Thus, our results show that the CSR3 and EcR3 enzymes were inhibited by distinct types of molecules, indicating that this HTS assay could be widely applied in drug discovery of class 1 RNase III enzymes.
更新日期:2020-05-21



























 京公网安备 11010802027423号
京公网安备 11010802027423号