当前位置:
X-MOL 学术
›
J. Mol. Cell. Cardiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A directed network analysis of the cardiome identifies molecular pathways contributing to the development of HFpEF.
Journal of Molecular and Cellular Cardiology ( IF 5 ) Pub Date : 2020-05-16 , DOI: 10.1016/j.yjmcc.2020.05.008 Georg Summer 1 , Annika R Kuhn 2 , Chantal Munts 2 , Daniela Miranda-Silva 3 , Adelino F Leite-Moreira 3 , André P Lourenço 3 , Stephane Heymans 4 , Inês Falcão-Pires 3 , Marc van Bilsen 2
Journal of Molecular and Cellular Cardiology ( IF 5 ) Pub Date : 2020-05-16 , DOI: 10.1016/j.yjmcc.2020.05.008 Georg Summer 1 , Annika R Kuhn 2 , Chantal Munts 2 , Daniela Miranda-Silva 3 , Adelino F Leite-Moreira 3 , André P Lourenço 3 , Stephane Heymans 4 , Inês Falcão-Pires 3 , Marc van Bilsen 2
Affiliation
|
AIMS
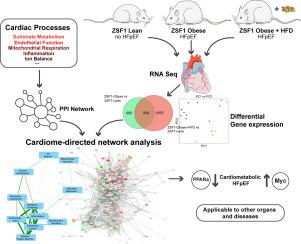
The metabolic syndrome and associated comorbidities, like diabetes, hypertension and obesity, have been implicated in the development of heart failure with preserved ejection fraction (HFpEF). The molecular mechanisms underlying the development of HFpEF remain to be elucidated. We developed a cardiome-directed network analysis and applied this to high throughput cardiac RNA-sequencing data from a well-established rat model of HFpEF, the obese and hypertensive ZSF1 rat. With this novel system biology approach, we explored the mechanisms underlying HFpEF.
METHODS AND RESULTS
Unlike ZSF1-Lean, ZSF1-Obese and ZSF1-Obese rats fed with a high-fat diet (HFD) developed diastolic dysfunction and reduced exercise capacity. The number of differentially expressed genes amounted to 1591 and 1961 for the ZSF1-Obese vs. Lean and ZSF1-Obese+HFD vs. Lean comparison, respectively. For the cardiome-directed network analysis (CDNA) eleven biological processes related to cardiac disease were selected and used as input for the STRING protein-protein interaction database. The resulting STRING network comprised 3.460 genes and 186.653 edges. Subsequently differentially expressed genes were projected onto this network. The connectivity between the core processes within the network was assessed and important bottleneck and hub genes were identified based on their network topology. Classical gene enrichment analysis highlighted many processes related to mitochondrial oxidative metabolism. The CDNA indicated high interconnectivity between five core processes: endothelial function, inflammation, apoptosis/autophagy, sarcomere/cytoskeleton and extracellular matrix. The transcription factors Myc and Peroxisome Proliferator-Activated Receptor-α (Ppara) were identified as important bottlenecks in the overall network topology, with Ppara acting as important link between cardiac metabolism, inflammation and endothelial function.
CONCLUSIONS
This study presents a novel systems biology approach, directly applicable to other cardiac disease-related transcriptome data sets. The CDNA approach enabled the identification of critical processes and genes, including Myc and Ppara, that are putatively involved in the development of HFpEF.
中文翻译:

对心脏的直接网络分析确定有助于HFpEF发展的分子途径。
目的代谢综合征和相关合并症,如糖尿病,高血压和肥胖症,与射血分数(HFpEF)维持在心力衰竭的发生有关。HFpEF发生的分子机制尚待阐明。我们开发了针对心脏的网络分析,并将其应用于来自成熟的HFpEF大鼠模型,肥胖和高血压ZSF1大鼠的高通量心脏RNA测序数据。通过这种新颖的系统生物学方法,我们探索了HFpEF的潜在机制。方法和结果与ZSF1-Lean不同,高脂饮食(HFD)喂养的ZSF1-Obese和ZSF1-Obese大鼠发展为舒张功能障碍并降低了运动能力。ZSF1-Obese vs.Lean和ZSF1-Obese + HFD vs.Zean的差异表达基因的数量分别为1591和1961。精益比较。对于心脏直接网络分析(CDNA),选择了11种与心脏病有关的生物学过程,并将其用作STRING蛋白质-蛋白质相互作用数据库的输入。所得的STRING网络包含3.460个基因和186.653个边缘。随后将差异表达的基因投影到该网络上。评估了网络内核心过程之间的连通性,并根据其网络拓扑确定了重要的瓶颈和枢纽基因。经典的基因富集分析突出了许多与线粒体氧化代谢有关的过程。CDNA表示五个核心过程之间的高度互连性:内皮功能,炎症,细胞凋亡/自噬,肌小节/细胞骨架和细胞外基质。转录因子Myc和过氧化物酶体增殖物激活的受体-α(Ppara)被确定为整个网络拓扑结构的重要瓶颈,Ppara成为心脏代谢,炎症和内皮功能之间的重要联系。结论本研究提出了一种新颖的系统生物学方法,可直接应用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。直接适用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。直接适用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。
更新日期:2020-05-16
中文翻译:

对心脏的直接网络分析确定有助于HFpEF发展的分子途径。
目的代谢综合征和相关合并症,如糖尿病,高血压和肥胖症,与射血分数(HFpEF)维持在心力衰竭的发生有关。HFpEF发生的分子机制尚待阐明。我们开发了针对心脏的网络分析,并将其应用于来自成熟的HFpEF大鼠模型,肥胖和高血压ZSF1大鼠的高通量心脏RNA测序数据。通过这种新颖的系统生物学方法,我们探索了HFpEF的潜在机制。方法和结果与ZSF1-Lean不同,高脂饮食(HFD)喂养的ZSF1-Obese和ZSF1-Obese大鼠发展为舒张功能障碍并降低了运动能力。ZSF1-Obese vs.Lean和ZSF1-Obese + HFD vs.Zean的差异表达基因的数量分别为1591和1961。精益比较。对于心脏直接网络分析(CDNA),选择了11种与心脏病有关的生物学过程,并将其用作STRING蛋白质-蛋白质相互作用数据库的输入。所得的STRING网络包含3.460个基因和186.653个边缘。随后将差异表达的基因投影到该网络上。评估了网络内核心过程之间的连通性,并根据其网络拓扑确定了重要的瓶颈和枢纽基因。经典的基因富集分析突出了许多与线粒体氧化代谢有关的过程。CDNA表示五个核心过程之间的高度互连性:内皮功能,炎症,细胞凋亡/自噬,肌小节/细胞骨架和细胞外基质。转录因子Myc和过氧化物酶体增殖物激活的受体-α(Ppara)被确定为整个网络拓扑结构的重要瓶颈,Ppara成为心脏代谢,炎症和内皮功能之间的重要联系。结论本研究提出了一种新颖的系统生物学方法,可直接应用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。直接适用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。直接适用于其他与心脏病有关的转录组数据集。CDNA方法可以鉴定关键过程和基因,包括Myc和Ppara,这些过程和基因可能与HFpEF的发展有关。



























 京公网安备 11010802027423号
京公网安备 11010802027423号