Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2020-05-15 , DOI: 10.1016/j.csbj.2020.05.002 Jorge Roel-Touris 1 , Alexandre M J J Bonvin 1
|
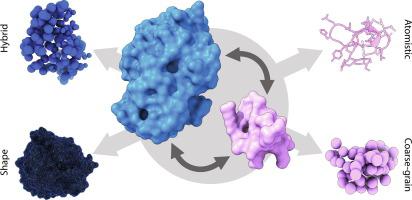
The computational modeling field has vastly evolved over the past decades. The early developments of simplified protein systems represented a stepping stone towards establishing more efficient approaches to sample intricated conformational landscapes. Downscaling the level of resolution of biomolecules to coarser representations allows for studying protein structure, dynamics and interactions that are not accessible by classical atomistic approaches. The combination of different resolutions, namely hybrid modeling, has also been proved as an alternative when mixed levels of details are required. In this review, we provide an overview of coarse-grained/hybrid models focusing on their applicability in the modeling of biomolecular interactions. We give a detailed list of ready-to-use modeling software for studying biomolecular interactions allowing various levels of coarse graining and provide examples of complexes determined by integrative coarse-grained/hybrid approaches in combination with experimental information.
中文翻译:

生物分子相互作用的粗粒度(混合)集成模型。
在过去的几十年中,计算建模领域发生了巨大的变化。简化蛋白质系统的早期发展代表了建立更有效的方法来采样复杂构象景观的踏脚石。将生物分子的分辨率降低到更粗略的表示形式,可以研究经典原子方法无法获得的蛋白质结构,动力学和相互作用。当需要混合的细节层次时,不同分辨率的组合即混合建模也已被证明是一种替代方法。在这篇综述中,我们提供了粗粒/混合模型的概述,重点是它们在生物分子相互作用建模中的适用性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号