当前位置:
X-MOL 学术
›
J. Neurosci. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
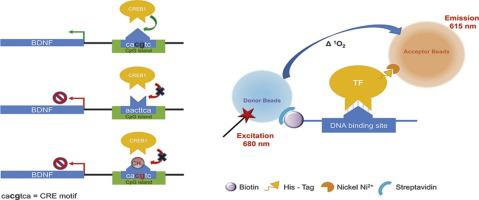
A new methodological approach for in vitro determination of the role of DNA methylation on transcription factor binding using AlphaScreen® analysis: Focus on CREB1 binding at hBDNF promoter IV.
Journal of Neuroscience Methods ( IF 3 ) Pub Date : 2020-05-13 , DOI: 10.1016/j.jneumeth.2020.108720 A Sabatucci 1 , V Berchet 2 , F Bellia 1 , M Maccarrone 3 , E Dainese 1 , C D'Addario 1 , M Pucci 1
Journal of Neuroscience Methods ( IF 3 ) Pub Date : 2020-05-13 , DOI: 10.1016/j.jneumeth.2020.108720 A Sabatucci 1 , V Berchet 2 , F Bellia 1 , M Maccarrone 3 , E Dainese 1 , C D'Addario 1 , M Pucci 1
Affiliation
|
BACKGROUND
DNA methylation plays a relevant role in the regulation of gene transcription, but currently the exact quantification of transcription factors binding to methylated DNA is not being determined. The binding of the transcription factor cAMP response element-binding protein-1 to its cognate CpG containing motif is known to be impaired upon methylation. It thus represents a paradigmatic system to experimentally verify the validity of a new in vitro method to measure the role of methylation on DNA/transcription factors binding.
METHOD
An AlphaScreen® assay was developed to quantitatively measure the contribution of DNA CpG methylation on the interaction with transcription factors. The method was validated measuring the variation in affinity of cAMP response element-binding protein-1 and its recognition motif in human Brain-derived neurotrophic factor gene exon IV promoter as a function of CpG methylation.
RESULTS
For the first time, a quantitative direct correlation between DNA methylation and transcription factors binding is reported showing a dramatic reduction in binding affinity between fully methylated and non-methylated DNA.
COMPARISON WITH EXISTING METHODS
This methodology allows to directly measure DNA/transcription factors binding ability as a function of DNA methylation levels thus improving not quantitative methods available today. Moreover, it allows to work with purified proteins and oligonucleotides without need of chromatin.
CONCLUSIONS
The present methodology is suggested as a new analytical tool for the quantitative determination of the effect of CpG methylation on the interaction of gene promoters with transcription factors regulating gene expression, a key epigenetic mechanism implicated in many physiological and pathological conditions.
中文翻译:

一种使用 AlphaScreen® 分析体外确定 DNA 甲基化对转录因子结合作用的新方法:关注 CREB1 在 hBDNF 启动子 IV 上的结合。
背景DNA甲基化在基因转录的调节中起相关作用,但目前尚未确定与甲基化DNA结合的转录因子的准确定量。已知转录因子 cAMP 反应元件结合蛋白-1 与其同源的含有 CpG 的基序的结合在甲基化时会受到损害。因此,它代表了一个范例系统,可以通过实验验证一种新的体外方法的有效性,以测量甲基化对 DNA/转录因子结合的作用。方法 开发了一种 AlphaScreen® 测定来定量测量 DNA CpG 甲基化对与转录因子相互作用的贡献。该方法通过测量 cAMP 反应元件结合蛋白-1 的亲和力变化及其在人脑源性神经营养因子基因外显子 IV 启动子中的识别基序随 CpG 甲基化的变化而得到验证。结果首次报道了DNA甲基化和转录因子结合之间的定量直接相关性,显示完全甲基化和非甲基化DNA之间的结合亲和力显着降低。与现有方法的比较该方法允许直接测量 DNA/转录因子结合能力作为 DNA 甲基化水平的函数,从而改进了当今可用的非定量方法。此外,它允许在不需要染色质的情况下处理纯化的蛋白质和寡核苷酸。
更新日期:2020-05-13
中文翻译:

一种使用 AlphaScreen® 分析体外确定 DNA 甲基化对转录因子结合作用的新方法:关注 CREB1 在 hBDNF 启动子 IV 上的结合。
背景DNA甲基化在基因转录的调节中起相关作用,但目前尚未确定与甲基化DNA结合的转录因子的准确定量。已知转录因子 cAMP 反应元件结合蛋白-1 与其同源的含有 CpG 的基序的结合在甲基化时会受到损害。因此,它代表了一个范例系统,可以通过实验验证一种新的体外方法的有效性,以测量甲基化对 DNA/转录因子结合的作用。方法 开发了一种 AlphaScreen® 测定来定量测量 DNA CpG 甲基化对与转录因子相互作用的贡献。该方法通过测量 cAMP 反应元件结合蛋白-1 的亲和力变化及其在人脑源性神经营养因子基因外显子 IV 启动子中的识别基序随 CpG 甲基化的变化而得到验证。结果首次报道了DNA甲基化和转录因子结合之间的定量直接相关性,显示完全甲基化和非甲基化DNA之间的结合亲和力显着降低。与现有方法的比较该方法允许直接测量 DNA/转录因子结合能力作为 DNA 甲基化水平的函数,从而改进了当今可用的非定量方法。此外,它允许在不需要染色质的情况下处理纯化的蛋白质和寡核苷酸。



























 京公网安备 11010802027423号
京公网安备 11010802027423号