当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Performance evaluation of cetacean species distribution models developed using generalized additive models and boosted regression trees.
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-05-11 , DOI: 10.1002/ece3.6316 Elizabeth A Becker 1, 2, 3 , James V Carretta 4 , Karin A Forney 5, 6 , Jay Barlow 4 , Stephanie Brodie 2, 7 , Ryan Hoopes 3 , Michael G Jacox 7, 8 , Sara M Maxwell 9 , Jessica V Redfern 10 , Nicholas B Sisson 11 , Heather Welch 2, 7 , Elliott L Hazen 2, 7
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-05-11 , DOI: 10.1002/ece3.6316 Elizabeth A Becker 1, 2, 3 , James V Carretta 4 , Karin A Forney 5, 6 , Jay Barlow 4 , Stephanie Brodie 2, 7 , Ryan Hoopes 3 , Michael G Jacox 7, 8 , Sara M Maxwell 9 , Jessica V Redfern 10 , Nicholas B Sisson 11 , Heather Welch 2, 7 , Elliott L Hazen 2, 7
Affiliation
|
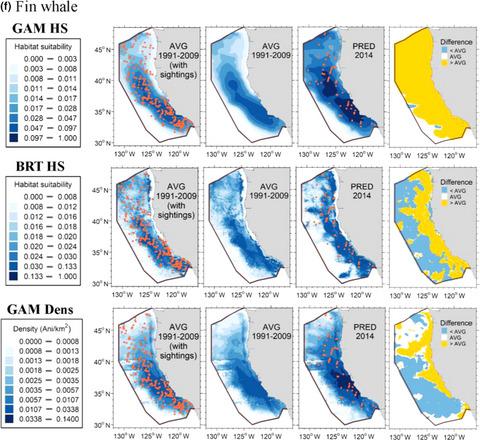
Species distribution models (SDMs) are important management tools for highly mobile marine species because they provide spatially and temporally explicit information on animal distribution. Two prevalent modeling frameworks used to develop SDMs for marine species are generalized additive models (GAMs) and boosted regression trees (BRTs), but comparative studies have rarely been conducted; most rely on presence‐only data; and few have explored how features such as species distribution characteristics affect model performance. Since the majority of marine species BRTs have been used to predict habitat suitability, we first compared BRTs to GAMs that used presence/absence as the response variable. We then compared results from these habitat suitability models to GAMs that predict species density (animals per km2) because density models built with a subset of the data used here have previously received extensive validation. We compared both the explanatory power (i.e., model goodness of fit) and predictive power (i.e., performance on a novel dataset) of the GAMs and BRTs for a taxonomically diverse suite of cetacean species using a robust set of systematic survey data (1991–2014) within the California Current Ecosystem. Both BRTs and GAMs were successful at describing overall distribution patterns throughout the study area for the majority of species considered, but when predicting on novel data, the density GAMs exhibited substantially greater predictive power than both the presence/absence GAMs and BRTs, likely due to both the different response variables and fitting algorithms. Our results provide an improved understanding of some of the strengths and limitations of models developed using these two methods. These results can be used by modelers developing SDMs and resource managers tasked with the spatial management of marine species to determine the best modeling technique for their question of interest.
中文翻译:

使用广义加性模型和增强回归树开发的鲸类物种分布模型的性能评估。
物种分布模型(SDM)是高度移动的海洋物种的重要管理工具,因为它们提供了有关动物分布的时空明确信息。用于开发海洋物种SDM的两个流行的建模框架是广义加性模型(GAM)和增强回归树(BRT),但是很少进行比较研究;这是很普遍的。大多数依赖于在线状态数据;很少有人探索诸如物种分布特征之类的特征如何影响模型性能。由于大多数海洋物种BRT已被用于预测栖息地的适宜性,因此我们首先将BRT与GAM进行了比较,GAM使用存在/不存在作为响应变量。然后,我们将这些栖息地适应性模型的结果与预测物种密度(每平方公里动物2),因为使用此处使用的数据子集构建的密度模型先前已得到广泛的验证。我们使用一套可靠的系统调查数据,比较了一组分类不同的鲸类物种的GAM和BRT的解释力(即模型拟合优度)和预测力(即新数据集的性能)。 2014年)。BRT和GAM都成功地描述了所考虑的大多数物种在整个研究区域的总体分布模式,但是当根据新数据进行预测时,密度GAM的预测能力远高于是否存在GAM和BRT,这可能是由于不同的响应变量和拟合算法。我们的结果提供了对使用这两种方法开发的模型的某些优势和局限性的更好理解。开发SDM的建模人员和负责海洋物种空间管理的资源管理人员可以使用这些结果来确定他们感兴趣的问题的最佳建模技术。
更新日期:2020-06-26
中文翻译:

使用广义加性模型和增强回归树开发的鲸类物种分布模型的性能评估。
物种分布模型(SDM)是高度移动的海洋物种的重要管理工具,因为它们提供了有关动物分布的时空明确信息。用于开发海洋物种SDM的两个流行的建模框架是广义加性模型(GAM)和增强回归树(BRT),但是很少进行比较研究;这是很普遍的。大多数依赖于在线状态数据;很少有人探索诸如物种分布特征之类的特征如何影响模型性能。由于大多数海洋物种BRT已被用于预测栖息地的适宜性,因此我们首先将BRT与GAM进行了比较,GAM使用存在/不存在作为响应变量。然后,我们将这些栖息地适应性模型的结果与预测物种密度(每平方公里动物2),因为使用此处使用的数据子集构建的密度模型先前已得到广泛的验证。我们使用一套可靠的系统调查数据,比较了一组分类不同的鲸类物种的GAM和BRT的解释力(即模型拟合优度)和预测力(即新数据集的性能)。 2014年)。BRT和GAM都成功地描述了所考虑的大多数物种在整个研究区域的总体分布模式,但是当根据新数据进行预测时,密度GAM的预测能力远高于是否存在GAM和BRT,这可能是由于不同的响应变量和拟合算法。我们的结果提供了对使用这两种方法开发的模型的某些优势和局限性的更好理解。开发SDM的建模人员和负责海洋物种空间管理的资源管理人员可以使用这些结果来确定他们感兴趣的问题的最佳建模技术。


























 京公网安备 11010802027423号
京公网安备 11010802027423号