当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Genotypic and phenotypic analyses reveal distinct population structures and ecotypes for sugar beet-associated Pseudomonas in Oxford and Auckland.
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-05-11 , DOI: 10.1002/ece3.6334 Xue-Xian Zhang 1, 2 , Stephen R Ritchie 1, 3 , Hao Chang 1 , Dawn L Arnold 4 , Robert W Jackson 5 , Paul B Rainey 1, 6, 7
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-05-11 , DOI: 10.1002/ece3.6334 Xue-Xian Zhang 1, 2 , Stephen R Ritchie 1, 3 , Hao Chang 1 , Dawn L Arnold 4 , Robert W Jackson 5 , Paul B Rainey 1, 6, 7
Affiliation
|
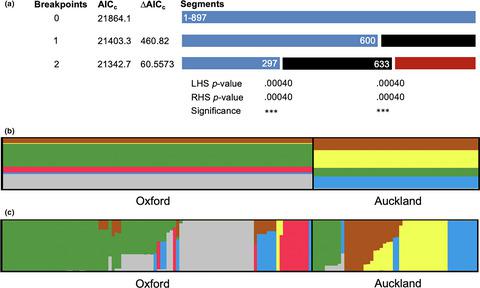
Fluorescent pseudomonads represent one of the largest groups of bacteria inhabiting the surfaces of plants, but their genetic composition in planta is poorly understood. Here, we examined the population structure and diversity of fluorescent pseudomonads isolated from sugar beet grown at two geographic locations (Oxford, United Kingdom and Auckland, New Zealand). To seek evidence for niche adaptation, bacteria were sampled from three types of leaves (immature, mature, and senescent) and then characterized using a combination of genotypic and phenotypic analysis. We first performed multilocus sequence analysis (MLSA) of three housekeeping genes (gapA , gltA , and acnB ) in a total of 152 isolates (96 from Oxford, 56 from Auckland). The concatenated sequences were grouped into 81 sequence types and 22 distinct operational taxonomic units (OTUs). Significant levels of recombination were detected, particularly for the Oxford isolates (rate of recombination to mutation (r/m) = 5.23 for the whole population). Subsequent ancestral analysis performed in STRUCTURE found evidence of six ancestral populations, and their distributions significantly differed between Oxford and Auckland. Next, their ability to grow on 95 carbon sources was assessed using the Biolog™ GN2 microtiter plates. A distance matrix was generated from the raw growth data (A 660) and subjected to multidimensional scaling (MDS) analysis. There was a significant correlation between substrate utilization profiles and MLSA genotypes. Both phenotypic and genotypic analyses indicated presence of a geographic structure for strains from Oxford and Auckland. Significant differences were also detected for MLSA genotypes between strains isolated from immature versus mature/senescent leaves. The fluorescent pseudomonads thus showed an ecotypic population structure, suggestive of adaptation to both geographic conditions and local plant niches.
中文翻译:

基因型和表型分析揭示了牛津和奥克兰的甜菜相关假单胞菌的不同种群结构和生态型。
荧光假单胞菌是居住在植物表面的最大细菌群之一,但人们对其在植物中的遗传组成了解甚少。在这里,我们研究了在两个地理位置(英国牛津和新西兰的奥克兰)种植的甜菜中分离出的荧光假单胞菌的种群结构和多样性。为了寻找适合生态位的证据,从三种类型的叶子(未成熟,成熟和衰老)中取样细菌,然后结合基因型和表型分析对其进行表征。我们首先对三个管家基因(gapA,gltA和acnB)进行了多基因座序列分析(MLSA)),共152株(牛津96株,奥克兰56株)。串联的序列分为81种序列类型和22种不同的操作分类单位(OTU)。检测到显着的重组水平,尤其是对于牛津分离株(整个人群的重组突变率(r / m)= 5.23)。随后在STRUCTURE中执行的祖先分析发现了六个祖先种群的证据,并且它们在牛津和奥克兰之间的分布存在显着差异。接下来,使用Biolog™GN2微量滴定板评估了它们在95个碳源上的生长能力。从原始生长数据生成距离矩阵(A 660),并进行了多维缩放(MDS)分析。底物利用率概况与MLSA基因型之间存在显着相关性。表型和基因型分析均表明存在来自牛津和奥克兰的菌株的地理结构。从未成熟叶与成熟/衰老叶分离的菌株之间,还发现了MLSA基因型的显着差异。因此,荧光假单胞菌显示出生态型种群结构,提示其既适应地理条件又适应本地植物生态位。
更新日期:2020-06-26
中文翻译:

基因型和表型分析揭示了牛津和奥克兰的甜菜相关假单胞菌的不同种群结构和生态型。
荧光假单胞菌是居住在植物表面的最大细菌群之一,但人们对其在植物中的遗传组成了解甚少。在这里,我们研究了在两个地理位置(英国牛津和新西兰的奥克兰)种植的甜菜中分离出的荧光假单胞菌的种群结构和多样性。为了寻找适合生态位的证据,从三种类型的叶子(未成熟,成熟和衰老)中取样细菌,然后结合基因型和表型分析对其进行表征。我们首先对三个管家基因(gapA,gltA和acnB)进行了多基因座序列分析(MLSA)),共152株(牛津96株,奥克兰56株)。串联的序列分为81种序列类型和22种不同的操作分类单位(OTU)。检测到显着的重组水平,尤其是对于牛津分离株(整个人群的重组突变率(r / m)= 5.23)。随后在STRUCTURE中执行的祖先分析发现了六个祖先种群的证据,并且它们在牛津和奥克兰之间的分布存在显着差异。接下来,使用Biolog™GN2微量滴定板评估了它们在95个碳源上的生长能力。从原始生长数据生成距离矩阵(A 660),并进行了多维缩放(MDS)分析。底物利用率概况与MLSA基因型之间存在显着相关性。表型和基因型分析均表明存在来自牛津和奥克兰的菌株的地理结构。从未成熟叶与成熟/衰老叶分离的菌株之间,还发现了MLSA基因型的显着差异。因此,荧光假单胞菌显示出生态型种群结构,提示其既适应地理条件又适应本地植物生态位。



























 京公网安备 11010802027423号
京公网安备 11010802027423号