当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Direct Sequencing of tRNA by 2D-HELS-AA MS Seq Reveals Its Different Isoforms and Dynamic Base Modifications.
ACS Chemical Biology ( IF 4 ) Pub Date : 2020-05-04 , DOI: 10.1021/acschembio.0c00119 Ning Zhang 1, 2 , Shundi Shi 2 , Xuanting Wang 2 , Wenhao Ni 1 , Xiaohong Yuan 1 , Jiachen Duan 1 , Tony Z Jia 3, 4 , Barney Yoo 5 , Ashley Ziegler 1 , James J Russo 2 , Wenjia Li 6 , Shenglong Zhang 1
ACS Chemical Biology ( IF 4 ) Pub Date : 2020-05-04 , DOI: 10.1021/acschembio.0c00119 Ning Zhang 1, 2 , Shundi Shi 2 , Xuanting Wang 2 , Wenhao Ni 1 , Xiaohong Yuan 1 , Jiachen Duan 1 , Tony Z Jia 3, 4 , Barney Yoo 5 , Ashley Ziegler 1 , James J Russo 2 , Wenjia Li 6 , Shenglong Zhang 1
Affiliation
|
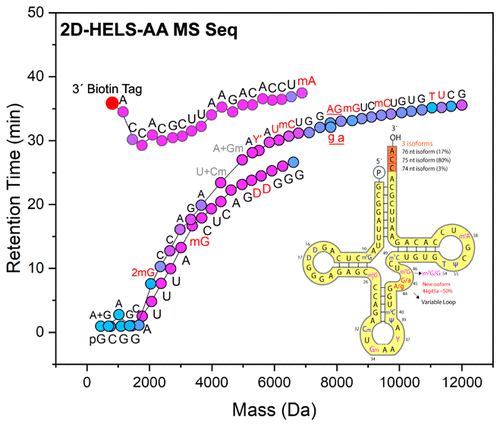
Post-transcriptional modifications are intrinsic to RNA structure and function. However, methods to sequence RNA typically require a cDNA intermediate and are either not able to sequence these modifications or are tailored to sequence one specific nucleotide modification only. Interestingly, some of these modifications occur with <100% frequency at their particular sites, and site-specific quantification of their stoichiometries is another challenge. Here, we report a direct method for sequencing tRNAPhe without cDNA by integrating a two-dimensional hydrophobic RNA end-labeling strategy with an anchor-based algorithm in mass spectrometry-based sequencing (2D-HELS-AA MS Seq). The entire tRNAPhe was sequenced and the identity, location, and stoichiometry of all eleven different RNA modifications was determined, five of which were not 100% modified, including a 2′-O-methylated G (Gm) in the wobble anticodon position as well as an N2, N2-dimethylguanosine (m22G), a 7-methylguanosine (m7G), a 1-methyladenosine (m1A), and a wybutosine (Y), suggesting numerous post-transcriptional regulations in tRNA. Two truncated isoforms at the 3′-CCA tail of the tRNAPhe (75 nt with a 3′-CC tail (80% abundance) and 74 nt with a 3′-C tail (3% abundance)) were identified in addition to the full-length 3′-CCA-tailed tRNAPhe (76 nt, 17% abundance). We discovered a new isoform with A–G transitions/editing at the 44 and 45 positions in the tRNAPhe variable loop, and discuss possible mechanisms related to the emergence and functions of the isoforms with these base transitions or editing. Our method revealed new isoforms, base modifications, and RNA editing as well as their stoichiometries in the tRNA that cannot be determined by current cDNA-based methods, opening new opportunities in the field of epitranscriptomics.
中文翻译:

通过2D-HELS-AA MS Seq直接测序tRNA揭示了其不同的同工型和动态碱基修饰。
转录后修饰是RNA结构和功能所固有的。但是,对RNA进行测序的方法通常需要cDNA中间体,或者不能对这些修饰进行测序,或者只能对一种特定的核苷酸修饰进行测序。有趣的是,这些修饰中的某些修饰在其特定位点以小于100%的频率发生,并且其化学计量的位点特异性定量是另一个挑战。在这里,我们报告了一种直接的方法,无需测序,即可在基于质谱的测序(2D-HELS-AA MS Seq)中将二维疏水性RNA末端标记策略与基于锚的算法整合在一起,从而无需cDNA即可对tRNA Phe进行测序。整个tRNA Phe测序并确定所有11种不同RNA修饰的身份,位置和化学计量,其中5种未100%修饰,包括摆动反密码子位置的2'-O-甲基化G(Gm)以及N在图2中,N 2-二甲基鸟苷(m 2 2 G),7-甲基鸟苷(m 7 G),1-甲基腺苷(m 1 A)和wybutosine(Y),提示tRNA中存在许多转录后调控。除了tRNA Phe的3'-CCA尾部(75 nt具有3'-CC尾部(80%丰度)和74 nt具有3'-C尾部(3%丰度)外,还鉴定出两个截短的同工型。全长3'-CCA尾tRNA Phe(76 nt,丰度为17%)。我们发现了一个新的同工型,在tRNA Phe可变环的44和45位具有A–G过渡/编辑,并讨论了与这些碱基过渡或编辑的同工型的出现和功能有关的可能机制。我们的方法揭示了新的同工型,碱基修饰,tRNA中的RNA编辑以及它们的化学计量,这些是目前基于cDNA的方法无法确定的,为表观转录组学领域提供了新的机会。
更新日期:2020-06-19
中文翻译:

通过2D-HELS-AA MS Seq直接测序tRNA揭示了其不同的同工型和动态碱基修饰。
转录后修饰是RNA结构和功能所固有的。但是,对RNA进行测序的方法通常需要cDNA中间体,或者不能对这些修饰进行测序,或者只能对一种特定的核苷酸修饰进行测序。有趣的是,这些修饰中的某些修饰在其特定位点以小于100%的频率发生,并且其化学计量的位点特异性定量是另一个挑战。在这里,我们报告了一种直接的方法,无需测序,即可在基于质谱的测序(2D-HELS-AA MS Seq)中将二维疏水性RNA末端标记策略与基于锚的算法整合在一起,从而无需cDNA即可对tRNA Phe进行测序。整个tRNA Phe测序并确定所有11种不同RNA修饰的身份,位置和化学计量,其中5种未100%修饰,包括摆动反密码子位置的2'-O-甲基化G(Gm)以及N在图2中,N 2-二甲基鸟苷(m 2 2 G),7-甲基鸟苷(m 7 G),1-甲基腺苷(m 1 A)和wybutosine(Y),提示tRNA中存在许多转录后调控。除了tRNA Phe的3'-CCA尾部(75 nt具有3'-CC尾部(80%丰度)和74 nt具有3'-C尾部(3%丰度)外,还鉴定出两个截短的同工型。全长3'-CCA尾tRNA Phe(76 nt,丰度为17%)。我们发现了一个新的同工型,在tRNA Phe可变环的44和45位具有A–G过渡/编辑,并讨论了与这些碱基过渡或编辑的同工型的出现和功能有关的可能机制。我们的方法揭示了新的同工型,碱基修饰,tRNA中的RNA编辑以及它们的化学计量,这些是目前基于cDNA的方法无法确定的,为表观转录组学领域提供了新的机会。


























 京公网安备 11010802027423号
京公网安备 11010802027423号