当前位置:
X-MOL 学术
›
J. Food Saf.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Analysis of the bacterial floral structure and diversity of Xuanwei ham by 16S rDNA sequencing
Journal of Food Safety ( IF 2.4 ) Pub Date : 2020-04-22 , DOI: 10.1111/jfs.12800 Luying Shan 1 , Yinjiao Li 1 , Shi Zheng 1 , Yuanmiao Wei 1 , Ying Shang 1, 2
Journal of Food Safety ( IF 2.4 ) Pub Date : 2020-04-22 , DOI: 10.1111/jfs.12800 Luying Shan 1 , Yinjiao Li 1 , Shi Zheng 1 , Yuanmiao Wei 1 , Ying Shang 1, 2
Affiliation
|
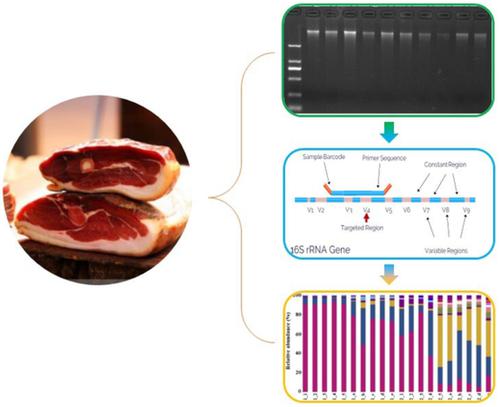
Xuanwei ham fermentation relies on indigenous and environmental microorganisms, which have abundant microbial resources. However, the traditional method of microbial community determination gives low detection rate of microorganisms. In order to reveal the microbial community of the Xuanwei ham more sufficiently, in this study, the total DNA (deoxyribonucleic acid) of the microbes from samples obtained from two brands of Xuanwei ham (10 samples each) were extracted with kit and SDS‐CTAB method. Afterward, 16S rDNA sequencing was performed to target the 16S rDNA gene V4 variant sequence. The composition and diversity of bacterial communities were analyzed. Results revealed that 872 operational taxonomic units, including 23 phyla, 39 classes, 61 orders, 131 families, and 258 genera, were obtained from the 20 ham samples at a similarity level of 97%. The bacterial diversity in Brand II was richer than that in Brand I. The DNA extracted by SDS‐CTAB could show the bacterial diversity better than that extracted by the kit method. At the genus level, the dominant bacterial communities were Tetragenococcus for Brand I and Acinetobacter for Brand II.
中文翻译:

用16S rDNA序列分析宣威火腿的细菌花序结构和多样性。
宣威火腿发酵依赖本地和环境微生物,这些微生物具有丰富的微生物资源。但是,传统的微生物群落测定方法对微生物的检测率较低。为了更充分地揭示宣威火腿的微生物群落,在本研究中,使用试剂盒和SDS-CTAB提取了来自两个宣威火腿品牌(每个样品10个)的样品中微生物的总DNA(脱氧核糖核酸)。方法。之后,进行16S rDNA测序以靶向16S rDNA基因V4变异序列。分析了细菌群落的组成和多样性。结果显示,从20个火腿样品中获得了872个可操作的生物分类单位,包括23个门,39个类,61个目,131个科和258个属,相似度为97%。品牌II中的细菌多样性要比品牌I中的丰富。通过SDS-CTAB提取的DNA可以显示出比试剂盒方法更好的细菌多样性。在属水平上,主要的细菌群落是品牌I为四球菌,品牌II为不动杆菌。
更新日期:2020-04-22
中文翻译:

用16S rDNA序列分析宣威火腿的细菌花序结构和多样性。
宣威火腿发酵依赖本地和环境微生物,这些微生物具有丰富的微生物资源。但是,传统的微生物群落测定方法对微生物的检测率较低。为了更充分地揭示宣威火腿的微生物群落,在本研究中,使用试剂盒和SDS-CTAB提取了来自两个宣威火腿品牌(每个样品10个)的样品中微生物的总DNA(脱氧核糖核酸)。方法。之后,进行16S rDNA测序以靶向16S rDNA基因V4变异序列。分析了细菌群落的组成和多样性。结果显示,从20个火腿样品中获得了872个可操作的生物分类单位,包括23个门,39个类,61个目,131个科和258个属,相似度为97%。品牌II中的细菌多样性要比品牌I中的丰富。通过SDS-CTAB提取的DNA可以显示出比试剂盒方法更好的细菌多样性。在属水平上,主要的细菌群落是品牌I为四球菌,品牌II为不动杆菌。



























 京公网安备 11010802027423号
京公网安备 11010802027423号