当前位置:
X-MOL 学术
›
Mol. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Parallel Generative Topographic Mapping: an Efficient Approach for Big Data Handling.
Molecular Informatics ( IF 3.6 ) Pub Date : 2020-04-29 , DOI: 10.1002/minf.202000009 Arkadii Lin 1 , Igor I Baskin 2 , Gilles Marcou 1 , Dragos Horvath 1 , Bernd Beck 3 , Alexandre Varnek 1
Molecular Informatics ( IF 3.6 ) Pub Date : 2020-04-29 , DOI: 10.1002/minf.202000009 Arkadii Lin 1 , Igor I Baskin 2 , Gilles Marcou 1 , Dragos Horvath 1 , Bernd Beck 3 , Alexandre Varnek 1
Affiliation
|
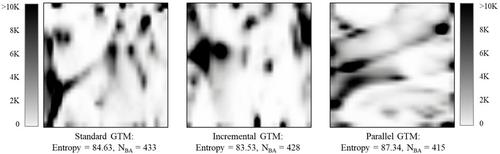
Generative Topographic Mapping (GTM) can be efficiently used to visualize, analyze and model large chemical data. The GTM manifold needs to span the chemical space deemed relevant for a given problem. Therefore, the Frame set (FS) of compounds used for the manifold construction must well cover a given chemical space. Intuitively, the FS size must raise with the size and diversity of the target library. At the same time, the GTM training can be very slow or even becomes technically impossible at FS sizes of the order of 105 compounds – which is a very small number compared to today's commercially accessible compounds, and, especially, to the theoretically feasible molecules. In order to solve this problem, we propose a Parallel GTM algorithm based on the merging of “intermediate” manifolds constructed in parallel for different subsets of molecules. An ensemble of these subsets forms a FS for the “final” manifold. In order to assess the efficiency of the new algorithm, 80 GTMs were built on the FSs of different sizes ranging from 10 to 1.8 M compounds selected from the ChEMBL database. Each GTM was challenged to build classification models for up to 712 biological activities (depending on the FS size). With the novel parallel GTM procedure, we could thus cover the entire spectrum of possible FS sizes, whereas previous studies were forced to rely on the working hypothesis that FS sizes of few thousands of compounds are sufficient to describe the ChEMBL chemical space. In fact, this study formally proves this to be true: a FS containing only 5000 randomly picked compounds is sufficient to represent the entire ChEMBL collection (1.8 M molecules), in the sense that a further increase of FS compound numbers has no benefice impact on the predictive propensity of the above‐mentioned 712 activity classification models. Parallel GTM may, however, be required to generate maps based on very large FS, that might improve chemical space cartography of big commercial and virtual libraries, approaching billions of compounds
中文翻译:

并行生成地形图:一种有效的大数据处理方法。
生成地形图 (GTM) 可有效地用于对大型化学数据进行可视化、分析和建模。GTM 流形需要跨越被认为与给定问题相关的化学空间。因此,用于歧管结构的化合物的框架集 (FS) 必须很好地覆盖给定的化学空间。直观地说,FS 大小必须随着目标库的大小和多样性而增加。同时,GTM 训练可能非常缓慢,甚至在 10 5数量级的 FS 大小时在技术上变得不可能化合物——与当今商业上可获得的化合物相比,尤其是与理论上可行的分子相比,这是一个非常小的数字。为了解决这个问题,我们提出了一种基于合并为不同分子子集并行构建的“中间”流形的并行 GTM 算法。这些子集的集合形成了“最终”流形的 FS。为了评估新算法的效率,在从 ChEMBL 数据库中选择的 10 到 1.8 M 化合物的不同大小的 FS 上构建了 80 个 GTM。每个 GTM 都面临着为多达 712 个生物活动(取决于 FS 大小)构建分类模型的挑战。通过新颖的并行 GTM 程序,我们可以覆盖可能的 FS 大小的整个范围,而之前的研究被迫依赖于工作假设,即数千种化合物的 FS 大小足以描述 ChEMBL 化学空间。事实上,这项研究正式证明了这一点:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),因为进一步增加 FS 化合物数量对上述 712 个活动分类模型的预测倾向。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物 这项研究正式证明这是真的:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),从某种意义上说,FS 化合物数量的进一步增加对预测倾向没有任何好处在上述 712 个活动分类模型中。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物 这项研究正式证明这是真的:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),从某种意义上说,FS 化合物数量的进一步增加对预测倾向没有任何好处在上述 712 个活动分类模型中。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物
更新日期:2020-04-29
中文翻译:

并行生成地形图:一种有效的大数据处理方法。
生成地形图 (GTM) 可有效地用于对大型化学数据进行可视化、分析和建模。GTM 流形需要跨越被认为与给定问题相关的化学空间。因此,用于歧管结构的化合物的框架集 (FS) 必须很好地覆盖给定的化学空间。直观地说,FS 大小必须随着目标库的大小和多样性而增加。同时,GTM 训练可能非常缓慢,甚至在 10 5数量级的 FS 大小时在技术上变得不可能化合物——与当今商业上可获得的化合物相比,尤其是与理论上可行的分子相比,这是一个非常小的数字。为了解决这个问题,我们提出了一种基于合并为不同分子子集并行构建的“中间”流形的并行 GTM 算法。这些子集的集合形成了“最终”流形的 FS。为了评估新算法的效率,在从 ChEMBL 数据库中选择的 10 到 1.8 M 化合物的不同大小的 FS 上构建了 80 个 GTM。每个 GTM 都面临着为多达 712 个生物活动(取决于 FS 大小)构建分类模型的挑战。通过新颖的并行 GTM 程序,我们可以覆盖可能的 FS 大小的整个范围,而之前的研究被迫依赖于工作假设,即数千种化合物的 FS 大小足以描述 ChEMBL 化学空间。事实上,这项研究正式证明了这一点:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),因为进一步增加 FS 化合物数量对上述 712 个活动分类模型的预测倾向。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物 这项研究正式证明这是真的:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),从某种意义上说,FS 化合物数量的进一步增加对预测倾向没有任何好处在上述 712 个活动分类模型中。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物 这项研究正式证明这是真的:仅包含 5000 个随机选择的化合物的 FS 足以代表整个 ChEMBL 集合(1.8 M 分子),从某种意义上说,FS 化合物数量的进一步增加对预测倾向没有任何好处在上述 712 个活动分类模型中。然而,可能需要并行 GTM 来生成基于非常大的 FS 的地图,这可能会改进大型商业和虚拟图书馆的化学空间制图,接近数十亿化合物


























 京公网安备 11010802027423号
京公网安备 11010802027423号