Journal of Pharmaceutical Analysis ( IF 8.8 ) Pub Date : 2020-04-28 , DOI: 10.1016/j.jpha.2020.04.008 Muhammad Usman Mirza 1 , Matheus Froeyen 1
|
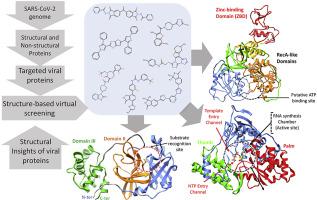
Recently emerged SARS-CoV-2 caused a major outbreak of coronavirus disease 2019 (COVID-19) and instigated a widespread fear, threatening global health safety. To date, no licensed antiviral drugs or vaccines are available against COVID-19 although several clinical trials are under way to test possible therapies. During this urgent situation, computational drug discovery methods provide an alternative to tiresome high-throughput screening, particularly in the hit-to-lead-optimization stage. Identification of small molecules that specifically target viral replication apparatus has indicated the highest potential towards antiviral drug discovery. In this work, we present potential compounds that specifically target SARS-CoV-2 vital proteins, including the main protease, Nsp12 RNA polymerase and Nsp13 helicase. An integrative virtual screening and molecular dynamics simulations approach has facilitated the identification of potential binding modes and favourable molecular interaction profile of corresponding compounds. Moreover, the identification of structurally important binding site residues in conserved motifs located inside the active site highlights relative importance of ligand binding based on residual energy decomposition analysis. Although the current study lacks experimental validation, the structural information obtained from this computational study has paved way for the design of targeted inhibitors to combat COVID-19 outbreak.
中文翻译:

SARS-CoV-2重要蛋白的结构阐明:计算方法揭示了针对主要蛋白酶,Nsp12聚合酶和Nsp13解旋酶的潜在候选药物。
最近出现的SARS-CoV-2引起了2019年冠状病毒病的重大爆发(COVID-19),并引发了广泛的恐惧,威胁了全球健康安全。迄今为止,尚无针对COVID-19的获得许可的抗病毒药物或疫苗,尽管正在进行数项临床试验以测试可能的疗法。在这种紧急情况下,计算药物发现方法为烦人的高通量筛选提供了一种替代方法,特别是在“铅到铅”优化阶段。鉴定特异性靶向病毒复制装置的小分子表明了抗病毒药物发现的最高潜力。在这项工作中,我们介绍了潜在地针对SARS-CoV-2重要蛋白的潜在化合物,包括主要蛋白酶,Nsp12 RNA聚合酶和Nsp13解旋酶。集成的虚拟筛选和分子动力学模拟方法促进了相应化合物潜在的结合模式的识别和有利的分子相互作用过程。此外,在位于活性位点内的保守基序中结构上重要的结合位点残基的鉴定突出了基于残余能量分解分析的配体结合的相对重要性。尽管当前的研究缺乏实验验证,但从该计算研究中获得的结构信息为抗击COVID-19爆发的靶向抑制剂的设计铺平了道路。在位于活性位点内的保守基序中结构上重要的结合位点残基的鉴定突出了基于残余能量分解分析的配体结合的相对重要性。尽管当前的研究缺乏实验验证,但从该计算研究中获得的结构信息为抗击COVID-19爆发的靶向抑制剂的设计铺平了道路。在位于活性位点内的保守基序中结构上重要的结合位点残基的鉴定突出了基于残余能量分解分析的配体结合的相对重要性。尽管当前的研究缺乏实验验证,但从该计算研究中获得的结构信息为抗击COVID-19爆发的靶向抑制剂的设计铺平了道路。


























 京公网安备 11010802027423号
京公网安备 11010802027423号