当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Protein–fragment complex structures derived by NMR molecular replacement
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-04-27 , DOI: 10.1039/d0md00068j Felix Torres 1, 2, 3, 4, 5 , Dhiman Ghosh 1, 2, 3, 4, 5 , Dean Strotz 1, 2, 3, 4, 5 , Celestine N. Chi 1, 2, 3, 4, 5 , Ben Davis 6, 7, 8 , Julien Orts 1, 2, 3, 4, 5
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-04-27 , DOI: 10.1039/d0md00068j Felix Torres 1, 2, 3, 4, 5 , Dhiman Ghosh 1, 2, 3, 4, 5 , Dean Strotz 1, 2, 3, 4, 5 , Celestine N. Chi 1, 2, 3, 4, 5 , Ben Davis 6, 7, 8 , Julien Orts 1, 2, 3, 4, 5
Affiliation
|
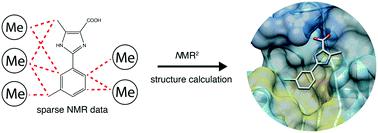
Recently we have established an NMR molecular replacement method, which is capable of solving the structure of the interaction site of protein–ligand complexes in a fully automated manner. While the method was successfully applied for ligands with strong and weak binding affinities, including small molecules and peptides, its applicability on ligand fragments remains to be shown. Structures of fragment–protein complexes are more challenging for the method since fragments contain only few protons. Here we show a successful application of the NMR molecular replacement method in solving structures of complexes between three derivatives of a ligand fragment and the protein receptor PIN1. We anticipate that this approach will find a broad application in fragment-based lead discovery.
中文翻译:

通过NMR分子置换得到的蛋白片段复杂结构
最近,我们建立了NMR分子置换方法,该方法能够以全自动方式解决蛋白质-配体复合物相互作用位点的结构。虽然该方法已成功应用于具有强和弱结合亲和力的配体,包括小分子和肽,但其在配体片段上的适用性仍有待证明。碎片-蛋白质复合物的结构对该方法更具挑战性,因为碎片仅包含少量质子。在这里,我们显示了NMR分子置换方法在解决配体片段的三个衍生物与蛋白质受体PIN1之间的复合物结构方面的成功应用。我们预计这种方法将在基于片段的线索发现中找到广泛的应用。
更新日期:2020-04-27
中文翻译:

通过NMR分子置换得到的蛋白片段复杂结构
最近,我们建立了NMR分子置换方法,该方法能够以全自动方式解决蛋白质-配体复合物相互作用位点的结构。虽然该方法已成功应用于具有强和弱结合亲和力的配体,包括小分子和肽,但其在配体片段上的适用性仍有待证明。碎片-蛋白质复合物的结构对该方法更具挑战性,因为碎片仅包含少量质子。在这里,我们显示了NMR分子置换方法在解决配体片段的三个衍生物与蛋白质受体PIN1之间的复合物结构方面的成功应用。我们预计这种方法将在基于片段的线索发现中找到广泛的应用。


























 京公网安备 11010802027423号
京公网安备 11010802027423号