Acta Pharmaceutica Sinica B ( IF 14.5 ) Pub Date : 2020-04-20 , DOI: 10.1016/j.apsb.2020.04.006 Yulong Shi 1, 2 , Xinben Zhang 1 , Kaijie Mu 1, 3 , Cheng Peng 1, 2 , Zhengdan Zhu 1, 2 , Xiaoyu Wang 1 , Yanqing Yang 1, 2 , Zhijian Xu 1, 2 , Weiliang Zhu 1, 2
|
Abstract
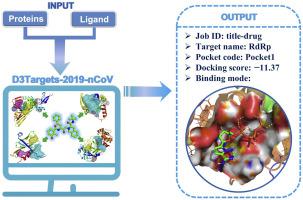
A highly effective medicine is urgently required to cure coronavirus disease 2019 (COVID-19). For the purpose, we developed a molecular docking based webserver, namely D3Targets-2019-nCoV, with two functions, one is for predicting drug targets for drugs or active compounds observed from clinic or in vitro/in vivo studies, the other is for identifying lead compounds against potential drug targets via docking. This server has its unique features, (1) the potential target proteins and their different conformations involving in the whole process from virus infection to replication and release were included as many as possible; (2) all the potential ligand-binding sites with volume larger than 200 Å3 on a protein structure were identified for docking; (3) correlation information among some conformations or binding sites was annotated; (4) it is easy to be updated, and is accessible freely to public (https://www.d3pharma.com/D3Targets-2019-nCoV/index.php). Currently, the webserver contains 42 proteins [20 severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) encoded proteins and 22 human proteins involved in virus infection, replication and release] with 69 different conformations/structures and 557 potential ligand-binding pockets in total. With 6 examples, we demonstrated that the webserver should be useful to medicinal chemists, pharmacologists and clinicians for efficiently discovering or developing effective drugs against the SARS-CoV-2 to cure COVID-19.
中文翻译:

D3Targets-2019-nCoV:用于预测药物靶点以及针对 COVID-19 进行多靶点和多站点虚拟筛选的网络服务器。
摘要
迫切需要一种高效的药物来治愈 2019 年冠状病毒病(COVID-19)。为此,我们开发了一种基于分子对接的网络服务器,即D3Targets-2019-nCoV,具有两个功能,一是预测临床或体外/体内研究中观察到的药物或活性化合物的药物靶点,二是识别通过对接引导化合物对抗潜在的药物靶点。该服务器有其独特之处,(1)尽可能多地包含了病毒从感染到复制释放全过程中潜在的目标蛋白及其不同构象;(2)鉴定蛋白质结构上所有体积大于200 Å 3的潜在配体结合位点并进行对接;(3)注释了一些构象或结合位点之间的相关信息;(4)易于更新,并且可供公众免费访问(https://www.d3pharma.com/D3Targets-2019-nCoV/index.php)。目前,该网络服务器包含 42 种蛋白质(20 种与严重急性呼吸综合征相关的冠状病毒 2(SARS-CoV-2)编码的蛋白质和 22 种参与病毒感染、复制和释放的人类蛋白质),具有 69 种不同的构象/结构和 557 种潜在配体 -总共装订口袋。我们通过 6 个例子证明,网络服务器应该对药物化学家、药理学家和临床医生有用,可以有效地发现或开发针对 SARS-CoV-2 的有效药物来治愈 COVID-19。


























 京公网安备 11010802027423号
京公网安备 11010802027423号