当前位置:
X-MOL 学术
›
Nat. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identifying genetic variants underlying phenotypic variation in plants without complete genomes.
Nature Genetics ( IF 30.8 ) Pub Date : 2020-04-13 , DOI: 10.1038/s41588-020-0612-7 Yoav Voichek 1 , Detlef Weigel 1
Nature Genetics ( IF 30.8 ) Pub Date : 2020-04-13 , DOI: 10.1038/s41588-020-0612-7 Yoav Voichek 1 , Detlef Weigel 1
Affiliation
|
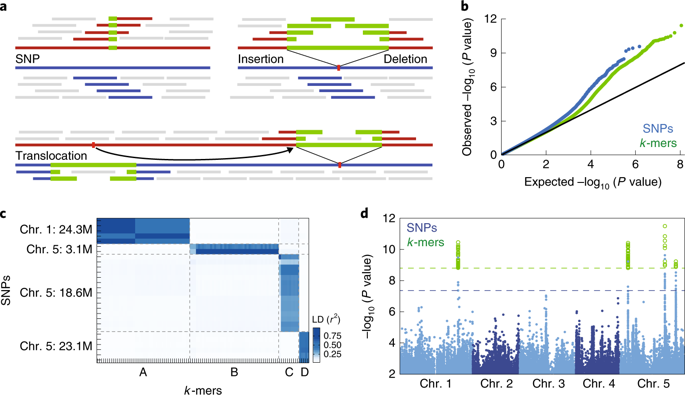
Structural variants and presence/absence polymorphisms are common in plant genomes, yet they are routinely overlooked in genome-wide association studies (GWAS). Here, we expand the type of genetic variants detected in GWAS to include major deletions, insertions and rearrangements. We first use raw sequencing data directly to derive short sequences, k-mers, that mark a broad range of polymorphisms independently of a reference genome. We then link k-mers associated with phenotypes to specific genomic regions. Using this approach, we reanalyzed 2,000 traits in Arabidopsis thaliana, tomato and maize populations. Associations identified with k-mers recapitulate those found with SNPs, but with stronger statistical support. Importantly, we discovered new associations with structural variants and with regions missing from reference genomes. Our results demonstrate the power of performing GWAS before linking sequence reads to specific genomic regions, which allows the detection of a wider range of genetic variants responsible for phenotypic variation.
中文翻译:

在没有完整基因组的情况下识别植物表型变异的遗传变异。
结构变异和存在/不存在多态性在植物基因组中很常见,但它们在全基因组关联研究 (GWAS) 中经常被忽视。在这里,我们扩展了 GWAS 中检测到的遗传变异类型,包括主要的缺失、插入和重排。我们首先直接使用原始测序数据来获得短序列,k-mers,它标志着独立于参考基因组的广泛多态性。然后,我们将与表型相关的 k-mer 与特定的基因组区域联系起来。使用这种方法,我们重新分析了拟南芥、番茄和玉米种群的 2,000 个性状。用 k-mer 确定的关联概括了用 SNP 发现的关联,但具有更强的统计支持。重要的是,我们发现了与结构变异和参考基因组中缺失区域的新关联。
更新日期:2020-04-24
中文翻译:

在没有完整基因组的情况下识别植物表型变异的遗传变异。
结构变异和存在/不存在多态性在植物基因组中很常见,但它们在全基因组关联研究 (GWAS) 中经常被忽视。在这里,我们扩展了 GWAS 中检测到的遗传变异类型,包括主要的缺失、插入和重排。我们首先直接使用原始测序数据来获得短序列,k-mers,它标志着独立于参考基因组的广泛多态性。然后,我们将与表型相关的 k-mer 与特定的基因组区域联系起来。使用这种方法,我们重新分析了拟南芥、番茄和玉米种群的 2,000 个性状。用 k-mer 确定的关联概括了用 SNP 发现的关联,但具有更强的统计支持。重要的是,我们发现了与结构变异和参考基因组中缺失区域的新关联。



























 京公网安备 11010802027423号
京公网安备 11010802027423号