当前位置:
X-MOL 学术
›
PLOS Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Inference of past demography, dormancy and self-fertilization rates from whole genome sequence data.
PLOS Genetics ( IF 4.5 ) Pub Date : 2020-04-06 , DOI: 10.1371/journal.pgen.1008698 Thibaut Paul Patrick Sellinger 1 , Diala Abu Awad 1 , Markus Moest 2 , Aurélien Tellier 1
PLOS Genetics ( IF 4.5 ) Pub Date : 2020-04-06 , DOI: 10.1371/journal.pgen.1008698 Thibaut Paul Patrick Sellinger 1 , Diala Abu Awad 1 , Markus Moest 2 , Aurélien Tellier 1
Affiliation
|
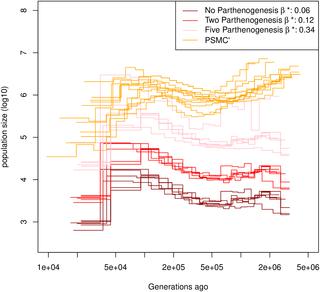
Several methods based on the Sequential Markovian coalescence (SMC) have been developed that make use of genome sequence data to uncover population demographic history, which is of interest in its own right and is a key requirement to generate a null model for selection tests. While these methods can be applied to all possible kind of species, the underlying assumptions are sexual reproduction in each generation and non-overlapping generations. However, in many plants, invertebrates, fungi and other taxa, those assumptions are often violated due to different ecological and life history traits, such as self-fertilization or long term dormant structures (seed or egg-banking). We develop a novel SMC-based method to infer 1) the rates/parameters of dormancy and of self-fertilization, and 2) the populations' past demographic history. Using simulated data sets, we demonstrate the accuracy of our method for a wide range of demographic scenarios and for sequence lengths from one to 30 Mb using four sampled genomes. Finally, we apply our method to a Swedish and a German population of Arabidopsis thaliana demonstrating a selfing rate of ca. 0.87 and the absence of any detectable seed-bank. In contrast, we show that the water flea Daphnia pulex exhibits a long lived egg-bank of three to 18 generations. In conclusion, we here present a novel method to infer accurate demographies and life-history traits for species with selfing and/or seed/egg-banks. Finally, we provide recommendations for the use of SMC-based methods for non-model organisms, highlighting the importance of the per site and the effective ratios of recombination over mutation.
中文翻译:

从全基因组序列数据推断过去的人口统计、休眠和自体受精率。
已经开发了几种基于顺序马尔可夫合并(SMC)的方法,这些方法利用基因组序列数据来揭示人口统计历史,这本身就很有趣,并且是生成用于选择测试的零模型的关键要求。虽然这些方法可以应用于所有可能的物种种类,但基本假设是每一代和非重叠世代的有性繁殖。然而,在许多植物、无脊椎动物、真菌和其他类群中,由于不同的生态和生活史特征,例如自体受精或长期休眠结构(种子或卵库),这些假设常常被违反。我们开发了一种基于 SMC 的新颖方法来推断 1) 休眠和自体受精的速率/参数,以及 2) 种群过去的人口统计历史。使用模拟数据集,我们使用四个采样基因组证明了我们的方法对于各种人口统计场景以及 1 到 30 Mb 序列长度的准确性。最后,我们将我们的方法应用于瑞典和德国的拟南芥群体,显示出约的自交率。0.87 并且不存在任何可检测到的种子库。相比之下,我们发现水蚤 Daphnia pulex 的卵库寿命较长,可达 3 至 18 代。总之,我们在这里提出了一种新方法来推断具有自交和/或种子/卵库的物种的准确人口统计学和生活史特征。最后,我们提供了针对非模式生物使用基于 SMC 的方法的建议,强调了每个位点的重要性以及重组与突变的有效比率。
更新日期:2020-04-06
中文翻译:

从全基因组序列数据推断过去的人口统计、休眠和自体受精率。
已经开发了几种基于顺序马尔可夫合并(SMC)的方法,这些方法利用基因组序列数据来揭示人口统计历史,这本身就很有趣,并且是生成用于选择测试的零模型的关键要求。虽然这些方法可以应用于所有可能的物种种类,但基本假设是每一代和非重叠世代的有性繁殖。然而,在许多植物、无脊椎动物、真菌和其他类群中,由于不同的生态和生活史特征,例如自体受精或长期休眠结构(种子或卵库),这些假设常常被违反。我们开发了一种基于 SMC 的新颖方法来推断 1) 休眠和自体受精的速率/参数,以及 2) 种群过去的人口统计历史。使用模拟数据集,我们使用四个采样基因组证明了我们的方法对于各种人口统计场景以及 1 到 30 Mb 序列长度的准确性。最后,我们将我们的方法应用于瑞典和德国的拟南芥群体,显示出约的自交率。0.87 并且不存在任何可检测到的种子库。相比之下,我们发现水蚤 Daphnia pulex 的卵库寿命较长,可达 3 至 18 代。总之,我们在这里提出了一种新方法来推断具有自交和/或种子/卵库的物种的准确人口统计学和生活史特征。最后,我们提供了针对非模式生物使用基于 SMC 的方法的建议,强调了每个位点的重要性以及重组与突变的有效比率。



























 京公网安备 11010802027423号
京公网安备 11010802027423号