当前位置:
X-MOL 学术
›
Sci. Total Environ.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Evaluation and sensitivity analysis of diatom DNA metabarcoding for WFD bioassessment of Mediterranean rivers.
Science of the Total Environment ( IF 9.8 ) Pub Date : 2020-04-04 , DOI: 10.1016/j.scitotenv.2020.138445 Javier Pérez-Burillo 1 , Rosa Trobajo 2 , Valentin Vasselon 3 , Frédéric Rimet 4 , Agnès Bouchez 4 , David G Mann 5
Science of the Total Environment ( IF 9.8 ) Pub Date : 2020-04-04 , DOI: 10.1016/j.scitotenv.2020.138445 Javier Pérez-Burillo 1 , Rosa Trobajo 2 , Valentin Vasselon 3 , Frédéric Rimet 4 , Agnès Bouchez 4 , David G Mann 5
Affiliation
|
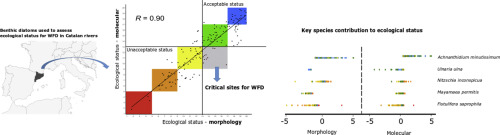
Our study of 164 diatom samples from Catalonia (NE Spain) is the first to evaluate the applicability of DNA metabarcoding, based on high throughput sequencing (HTS) using a 312-bp rbcL marker, for biomonitoring Mediterranean rivers. For this, we compared the values of a biotic index (IPS) and the ecological status classes derived from them, between light microscope-based (LM) and HTS methods. Very good correspondence between methods gives encouraging results concerning the applicability of DNA metabarcoding for Catalan rivers for the EU Water Framework Directive (WFD). However, in 10 sites, the ecological status class was downgraded from "Good"/"High" obtained by LM to "Moderate"/"Poor"/"Bad" by HTS; these "critical" sites are especially important, because the WFD requires remedial action by water managers for any river with Moderate or lower status. We investigated the contribution of each species to the IPS using a "leave-one-out" sensitivity analysis, paying special attention to critical sites. Discrepancies in IPS between LM and HTS were mainly due to the misidentification and overlooking in LM of a few species, which were better recovered by HTS. This bias was particularly important in the case of Fistulifera saprophila, whose clear underrepresentation in LM was important for explaining 8 out of the 10 critical sites and probably reflected destruction of weakly-silicified frustules during sample preparation. Differences between species in the rbcL copy number per cell affected the relative abundance obtained by HTS for Achnanthidium minutissimum, Nitzschia inconspicua and Ulnaria ulna, which were also identified by the sensitivity analysis as important for the WFD. Only minor IPS discrepancies were attributed to the incompleteness of the reference library, as most of the abundant and influential species (to the IPS) were well represented there. Finally, we propose that leave-one-out analysis is a good method for identifying priority species for isolation and barcoding.
中文翻译:

硅藻DNA元条形码对地中海河流WFD生物评估的评估和敏感性分析。
我们对来自加泰罗尼亚(西班牙东北部)的164个硅藻样品的研究是第一个基于高通量测序(HTS)(使用312-bp rbcL标记)评估DNA元条形码的适用性的生物监测地中海河流。为此,我们比较了基于光学显微镜(LM)和HTS方法的生物指数(IPS)值和从中得出的生态状况等级。方法之间的良好对应性给出了令人鼓舞的结果,涉及DNA加条形码在加泰罗尼亚河上的适用于欧盟水框架指令(WFD)。但是,在10个地点中,生态状况等级从LM获得的“好” /“高”降为HTS获得的“中等” /“差” /“差”。这些“关键”站点尤其重要,因为WFD要求水管理人员对状态为中度或较低状态的任何河流采取补救措施。我们使用“留一法”敏感性分析调查了每个物种对IPS的贡献,并特别注意了关键部位。LM和HTS之间IPS的差异主要是由于少数物种在LM中的错误识别和忽略,而HTS可以更好地弥补它们。这种偏见在腐生油菜(Fistulifera saprophila)的情况下尤为重要,在LM中明显的代表性不足对于解释10个关键位点中的8个很重要,并且很可能反映了样品制备过程中弱硅化的壳层的破坏。每个细胞的rbcL拷贝数物种之间的差异影响了HTS对min虫的相对丰度,敏感度分析还确定了Nitzschia inconspicua和Ulnaria尺骨对WFD很重要。只有少量的IPS差异归因于参考库的不完整,因为大多数丰富和有影响力的物种(对IPS而言)都可以很好地表示出来。最后,我们提出留一法分析是一种识别优先种类以进行隔离和条形码的好方法。
更新日期:2020-04-06
中文翻译:

硅藻DNA元条形码对地中海河流WFD生物评估的评估和敏感性分析。
我们对来自加泰罗尼亚(西班牙东北部)的164个硅藻样品的研究是第一个基于高通量测序(HTS)(使用312-bp rbcL标记)评估DNA元条形码的适用性的生物监测地中海河流。为此,我们比较了基于光学显微镜(LM)和HTS方法的生物指数(IPS)值和从中得出的生态状况等级。方法之间的良好对应性给出了令人鼓舞的结果,涉及DNA加条形码在加泰罗尼亚河上的适用于欧盟水框架指令(WFD)。但是,在10个地点中,生态状况等级从LM获得的“好” /“高”降为HTS获得的“中等” /“差” /“差”。这些“关键”站点尤其重要,因为WFD要求水管理人员对状态为中度或较低状态的任何河流采取补救措施。我们使用“留一法”敏感性分析调查了每个物种对IPS的贡献,并特别注意了关键部位。LM和HTS之间IPS的差异主要是由于少数物种在LM中的错误识别和忽略,而HTS可以更好地弥补它们。这种偏见在腐生油菜(Fistulifera saprophila)的情况下尤为重要,在LM中明显的代表性不足对于解释10个关键位点中的8个很重要,并且很可能反映了样品制备过程中弱硅化的壳层的破坏。每个细胞的rbcL拷贝数物种之间的差异影响了HTS对min虫的相对丰度,敏感度分析还确定了Nitzschia inconspicua和Ulnaria尺骨对WFD很重要。只有少量的IPS差异归因于参考库的不完整,因为大多数丰富和有影响力的物种(对IPS而言)都可以很好地表示出来。最后,我们提出留一法分析是一种识别优先种类以进行隔离和条形码的好方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号