当前位置:
X-MOL 学术
›
Mol. Phylogenet. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Lost in translation: Population genomics and long-read sequencing reveals relaxation of concerted evolution of the ribosomal DNA cistron.
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2020-04-02 , DOI: 10.1016/j.ympev.2020.106804 Keaton Tremble 1 , Laura M Suz 2 , Bryn T M Dentinger 1
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2020-04-02 , DOI: 10.1016/j.ympev.2020.106804 Keaton Tremble 1 , Laura M Suz 2 , Bryn T M Dentinger 1
Affiliation
|
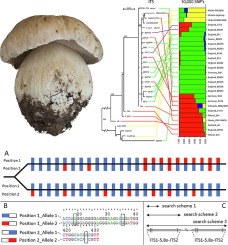
Concerted evolution of the ribosomal DNA array has been studied in numerous eukaryotic taxa, yet is still poorly understood. rDNA genes are repeated dozens to hundreds of times in the eukaryotic genome (Eickbush and Eickbush, 2007) and it is believed that these arrays are homogenized through concerted evolution (Zimmer et al., 1980; Dover, 1993) preventing the accumulation of intragenomic, and intraspecific, variation. However, numerous studies have reported rampant intragenomic and intraspecific variation in the rDNA array (Ganley and Kobayashi, 2011; Naidoo et al., 2013; Hughes and Petersen, 2001; Lindner and Banik, 2011; Li et al., 2013; Lindner et al., 2013; Hughes et al., 2018), contradicting our current understanding of concerted evolution. The internal transcribed spacers (ITS) of the rDNA cistron are the most commonly used DNA barcoding region in Fungi (Schoch et al., 2012), and rely on concerted evolution to homogenize the rDNA array leading to a "barcode gap" (Puillandre et al., 2012). Here we show that in Boletus edulis Bull., ITS intragenomic variation persists at low allele frequencies throughout the rDNA array, this variation does not correlate with genomic relatedness between populations, and rDNA genes may not evolve in a strictly concerted fashion despite the presence of unequal recombination and gene conversion. Under normal assumptions, heterozygous positions found in ITS sequences represent hybridization between populations, yet through allelic mapping of the rDNA array we found numerous heterozygous alleles to be stochastically introgressed throughout, presenting a dishonest signal of gene flow. Moreover, despite the signal of gene flow in ITS, our organisms were highly inbred, indicating a disconnect between true gene flow and barcoding signals. In addition, we show that while the mechanisms of concerted evolution are ongoing in pseudo-heterozygous individuals, they are not fully homogenizing the ITS array. Concerted evolution of the rDNA array may insufficiently homogenize the ITS gene, allowing for misleading signals of gene flow to persist, vastly complicating the use of the ITS locus for DNA barcoding in Fungi.
中文翻译:

翻译中遗失:种群基因组学和长期阅读测序揭示了核糖体DNA顺反子协同进化的松弛。
核糖体DNA阵列的协调进化已在许多真核生物类群中进行了研究,但仍知之甚少。rDNA基因在真核基因组中重复了数十到数百次(Eickbush和Eickbush,2007年),据信这些阵列通过协调一致的进化而被均质化(Zimmer等人,1980年; Dover,1993年),阻止了内部基因组的积累,以及种内变异。然而,许多研究报告说rDNA阵列中的基因组内和种内变异非常普遍(Ganley和Kobayashi,2011; Naidoo等,2013; Hughes和Petersen,2001; Lindner和Banik,2011; Li等,2013; Lindner等)等人,2013年;休斯等人,2018年),这与我们目前对协调进化的理解相矛盾。rDNA顺反子的内部转录间隔子(ITS)是真菌中最常用的DNA条形码区域(Schoch et al。,2012),并依靠协同进化来使rDNA阵列均质化,从而导致“条形码缺口”(Puillandre等)等人,2012年)。在这里,我们发现在牛肝菌中,ITS基因组内变异始终在低等位基因频率下在整个rDNA阵列中持续存在,这种变异与种群之间的基因组相关性不相关,尽管存在不相等的现象,rDNA基因也可能不会以严格一致的方式进化重组和基因转化。在正常的假设下,在ITS序列中发现的杂合子位置代表种群之间的杂交,但是通过rDNA阵列的等位基因作图,我们发现许多杂合子等位基因在整个过程中都是随机渗入的,呈现不真实的基因流信号。此外,尽管ITS中存在基因流信号,但我们的生物体高度近交,表明真实基因流与条形码信号之间存在脱节。此外,我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。
更新日期:2020-04-03
中文翻译:

翻译中遗失:种群基因组学和长期阅读测序揭示了核糖体DNA顺反子协同进化的松弛。
核糖体DNA阵列的协调进化已在许多真核生物类群中进行了研究,但仍知之甚少。rDNA基因在真核基因组中重复了数十到数百次(Eickbush和Eickbush,2007年),据信这些阵列通过协调一致的进化而被均质化(Zimmer等人,1980年; Dover,1993年),阻止了内部基因组的积累,以及种内变异。然而,许多研究报告说rDNA阵列中的基因组内和种内变异非常普遍(Ganley和Kobayashi,2011; Naidoo等,2013; Hughes和Petersen,2001; Lindner和Banik,2011; Li等,2013; Lindner等)等人,2013年;休斯等人,2018年),这与我们目前对协调进化的理解相矛盾。rDNA顺反子的内部转录间隔子(ITS)是真菌中最常用的DNA条形码区域(Schoch et al。,2012),并依靠协同进化来使rDNA阵列均质化,从而导致“条形码缺口”(Puillandre等)等人,2012年)。在这里,我们发现在牛肝菌中,ITS基因组内变异始终在低等位基因频率下在整个rDNA阵列中持续存在,这种变异与种群之间的基因组相关性不相关,尽管存在不相等的现象,rDNA基因也可能不会以严格一致的方式进化重组和基因转化。在正常的假设下,在ITS序列中发现的杂合子位置代表种群之间的杂交,但是通过rDNA阵列的等位基因作图,我们发现许多杂合子等位基因在整个过程中都是随机渗入的,呈现不真实的基因流信号。此外,尽管ITS中存在基因流信号,但我们的生物体高度近交,表明真实基因流与条形码信号之间存在脱节。此外,我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。我们显示,虽然伪杂合子个体正在进行协调进化的机制,但它们并未完全均化ITS阵列。rDNA阵列的协调进化可能不足以使ITS基因同质化,从而使基因流的误导信号持续存在,这极大地增加了将ITS基因座用于真菌中DNA条形码的复杂性。

























 京公网安备 11010802027423号
京公网安备 11010802027423号