当前位置:
X-MOL 学术
›
Comp. Biochem. Physiol. B Biochem. Mol. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification of differentially expressed genes in gills of tiger puffer (Takifugu rubripes) in response to low-salinity stress.
Comparative Biochemistry and Physiology B: Biochemistry & Molecular Biology ( IF 2.2 ) Pub Date : 2020-04-01 , DOI: 10.1016/j.cbpb.2020.110437 Jie-Lan Jiang 1 , Jia Xu 1 , Lin Ye 1 , Meng-Lei Sun 1 , Zhi-Qiang Jiang 1 , Ming-Guang Mao 1
Comparative Biochemistry and Physiology B: Biochemistry & Molecular Biology ( IF 2.2 ) Pub Date : 2020-04-01 , DOI: 10.1016/j.cbpb.2020.110437 Jie-Lan Jiang 1 , Jia Xu 1 , Lin Ye 1 , Meng-Lei Sun 1 , Zhi-Qiang Jiang 1 , Ming-Guang Mao 1
Affiliation
|
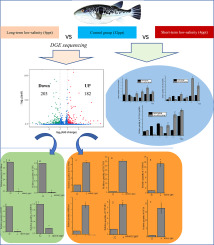
Salinity is an important abiotic factor for aquatic organisms. In fish, changes in salinity affect physiological responses and alter the immune system. Takifugu rubripes is an important economic marine fish, and mechanisms of T. rubripes adaptation to salinity changes need to be further documented. In this study, a transcriptome sequencing technique was used to analyse genes that were differentially expressed in the T. rubripes gill after low-salinity stress for 30 d, and differential gene expression was further validated by quantitative real-time PCR (qPCR). After assembly, 385 differentially expressed genes (DEGs) were identified, including 182 upregulated genes and 203 downregulated genes. The DEGs were assigned to Gene Ontology (GO) classes with a total of 1647 functional terms. Most DEGs were assigned to biological process (984; 59.8%) followed by molecular function (445; 27.0%) and cellular component (218; 13.2%). Further KEGG analysis allocated 385 DEGs to 95 KEGG pathways. After q-value correction, 7 pathways (Glycolysis/Gluconeogenesis; Biosynthesis of amino acids; Carbon metabolism; Fructose and mannose metabolism; Pentose phosphate pathway; Metabolism of xenobiotics by cytochrome P450; and Glycine, serine and threonine metabolism) remained significant. qPCR results indicated that the transcripts of six selected genes sharply increased after 30 d of low-salinity stress. Low-salinity stress obviously increased SLC39A6, SLC5A9, NKAα1, CYP1A1, CYP1B1, and GSTA expression. In contrast, the genes encoding Aldoaa, GPI, FBP2 and GAPDH exhibited downregulation. In addition, three solute carrier (SLC) genes selected from the DEGs were further studied for differential expression patterns after low-salinity exposure, and the results showed that the SLCs were upregulated in T. rubripes after 72 h of low-salinity exposure. This investigation provides data for understanding the molecular mechanisms of fish responses to low-salinity stress and provides a reference for rationally setting salinity levels in aquaculture.
中文翻译:

鉴定低盐胁迫下虎tiger(Takifugu rubripes)g中差异表达的基因。
盐度是水生生物的重要非生物因子。在鱼类中,盐度的变化会影响生理反应并改变免疫系统。东方红鱼是重要的经济海水鱼类,东方红T鱼适应盐度变化的机制有待进一步证明。在这项研究中,转录组测序技术用于分析低盐度胁迫30 d后在红唇T中差异表达的基因,并通过定量实时PCR(qPCR)进一步验证了差异基因的表达。组装后,鉴定了385个差异表达基因(DEG),包括182个上调基因和203个下调基因。将DEG分配给具有总共1647个功能术语的基因本体论(GO)类。大多数DEG被分配到生物过程中(984; 59。8%),其次是分子功能(445; 27.0%)和细胞成分(218; 13.2%)。进一步的KEGG分析将385个DEG分配给95个KEGG途径。在q值校正后,7个途径(糖酵解/糖异生;氨基酸的生物合成;碳代谢;果糖和甘露糖代谢;磷酸戊糖途径;细胞色素P450异源生物的代谢;甘氨酸,丝氨酸和苏氨酸的代谢)仍然很重要。qPCR结果表明,低盐胁迫30 d后,六个选定基因的转录本急剧增加。低盐度胁迫明显增加了SLC39A6,SLC5A9,NKAα1,CYP1A1,CYP1B1和GSTA的表达。相反,编码Aldoaa,GPI,FBP2和GAPDH的基因表现出下调。此外,进一步研究了从DEGs中选出的3个溶质载体(SLC)基因在低盐度暴露后的差异表达模式,结果表明,低盐度暴露72 h后,红豆中的SLCs被上调。这项调查提供了数据,以了解鱼类对低盐度胁迫的反应的分子机制,并为合理设定水产养殖中的盐度水平提供参考。
更新日期:2020-04-01
中文翻译:

鉴定低盐胁迫下虎tiger(Takifugu rubripes)g中差异表达的基因。
盐度是水生生物的重要非生物因子。在鱼类中,盐度的变化会影响生理反应并改变免疫系统。东方红鱼是重要的经济海水鱼类,东方红T鱼适应盐度变化的机制有待进一步证明。在这项研究中,转录组测序技术用于分析低盐度胁迫30 d后在红唇T中差异表达的基因,并通过定量实时PCR(qPCR)进一步验证了差异基因的表达。组装后,鉴定了385个差异表达基因(DEG),包括182个上调基因和203个下调基因。将DEG分配给具有总共1647个功能术语的基因本体论(GO)类。大多数DEG被分配到生物过程中(984; 59。8%),其次是分子功能(445; 27.0%)和细胞成分(218; 13.2%)。进一步的KEGG分析将385个DEG分配给95个KEGG途径。在q值校正后,7个途径(糖酵解/糖异生;氨基酸的生物合成;碳代谢;果糖和甘露糖代谢;磷酸戊糖途径;细胞色素P450异源生物的代谢;甘氨酸,丝氨酸和苏氨酸的代谢)仍然很重要。qPCR结果表明,低盐胁迫30 d后,六个选定基因的转录本急剧增加。低盐度胁迫明显增加了SLC39A6,SLC5A9,NKAα1,CYP1A1,CYP1B1和GSTA的表达。相反,编码Aldoaa,GPI,FBP2和GAPDH的基因表现出下调。此外,进一步研究了从DEGs中选出的3个溶质载体(SLC)基因在低盐度暴露后的差异表达模式,结果表明,低盐度暴露72 h后,红豆中的SLCs被上调。这项调查提供了数据,以了解鱼类对低盐度胁迫的反应的分子机制,并为合理设定水产养殖中的盐度水平提供参考。


























 京公网安备 11010802027423号
京公网安备 11010802027423号