当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Probing CRISPR-Cas12a Nuclease Activity Using Double-Stranded DNA-Templated Fluorescent Substrates.
Biochemistry ( IF 2.9 ) Pub Date : 2020-04-07 , DOI: 10.1021/acs.biochem.0c00140 Christopher W Smith 1 , Nidhi Nandu 1 , Mahera J Kachwala 1 , Yu-Sheng Chen 1 , Taha Bilal Uyar 1 , Mehmet V Yigit 1, 2
Biochemistry ( IF 2.9 ) Pub Date : 2020-04-07 , DOI: 10.1021/acs.biochem.0c00140 Christopher W Smith 1 , Nidhi Nandu 1 , Mahera J Kachwala 1 , Yu-Sheng Chen 1 , Taha Bilal Uyar 1 , Mehmet V Yigit 1, 2
Affiliation
|
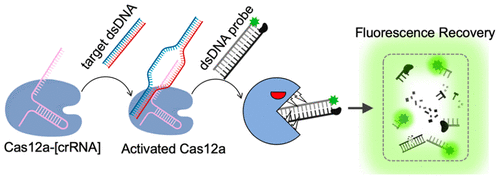
The CRISPR-Cas12a nuclease shreds short single-stranded DNA (ssDNA) substrates indiscriminately through trans-cleavage upon activation with a specific target DNA. This shredding activity offered the potential for development of ssDNA-templated probes with fluorescent dye (F) and quencher (Q) labels. However, the formulations of double-stranded DNA (dsDNA)-templated fluorescent probes have not been reported possibly due to unknown (or limited) activity of Cas12a against short dsDNAs. The ssDNA probes have been shown to be powerful for diagnostic applications; however, limiting the probe selections to short ssDNAs could be restrictive from an application and probe diversification standpoint. Here, we report a dsDNA substrate (probe-full) for probing Cas12a trans-cleavage activity upon target detection. A diverse set of Cas12a substrates with alternating dsDNA character were designed and studied using fluorescence spectroscopy. We have observed that probe-full without any nick displayed trans-cleavage performance that was better than that of the form that contains a nick. Different experimental conditions of salt concentration, target concentration, and mismatch tolerance were examined to evaluate the probe performance. The activity of Cas12a was programmed for a dsDNA frame copied from a tobacco curly shoot virus (TCSV) or hepatitis B virus (HepBV) genome by using crRNA against TCSV or HepBV, respectively. While on-target activity offered detection of as little as 10 pM dsDNA target, off-target activity was not observed even at 1 nM control DNAs. This study demonstrates that trans-cleavage of Cas12a is not limited to ssDNA substrates, and Cas12a-based diagnostics can be extended to dsDNA substrates.
中文翻译:

使用双链DNA模板荧光底物探测CRISPR-Cas12a核酸酶活性。
CRISPR-Cas12a核酸酶通过特异性靶DNA激活后进行反式切割,可随意切碎短的单链DNA(ssDNA)底物。这种切碎活性为开发带有荧光染料(F)和淬灭剂(Q)标记的ssDNA模板的探针提供了潜力。但是,由于Cas12a对短dsDNA的活性未知(或有限),因此尚未报道以双链DNA(dsDNA)为模板的荧光探针的配方。ssDNA探针已被证明在诊断应用中功能强大。但是,从应用和探针多样化的角度来看,将探针的选择限制在短的ssDNA上可能会受到限制。在这里,我们报告了一个双链DNA底物(充满探针),用于探测目标检测后的Cas12a反式切割活性。使用荧光光谱法设计和研究了具有交替dsDNA特性的多种Cas12a底物。我们已经观察到没有任何切口的充满探针的探针显示的反切割性能要好于包含切口的形式。检查盐浓度,目标浓度和不匹配耐受性的不同实验条件,以评估探针性能。通过分别使用针对TCSV或HepBV的crRNA,针对从烟草卷曲枝病毒(TCSV)或乙型肝炎病毒(HepBV)基因组复制的dsDNA框架对Cas12a的活性进行编程。虽然按靶标活性可检测到低至10 pM dsDNA靶标,但即使在1 nM对照DNA上也未观察到离靶标活性。这项研究表明,Cas12a的反式切割不仅限于ssDNA底物,
更新日期:2020-04-23
中文翻译:

使用双链DNA模板荧光底物探测CRISPR-Cas12a核酸酶活性。
CRISPR-Cas12a核酸酶通过特异性靶DNA激活后进行反式切割,可随意切碎短的单链DNA(ssDNA)底物。这种切碎活性为开发带有荧光染料(F)和淬灭剂(Q)标记的ssDNA模板的探针提供了潜力。但是,由于Cas12a对短dsDNA的活性未知(或有限),因此尚未报道以双链DNA(dsDNA)为模板的荧光探针的配方。ssDNA探针已被证明在诊断应用中功能强大。但是,从应用和探针多样化的角度来看,将探针的选择限制在短的ssDNA上可能会受到限制。在这里,我们报告了一个双链DNA底物(充满探针),用于探测目标检测后的Cas12a反式切割活性。使用荧光光谱法设计和研究了具有交替dsDNA特性的多种Cas12a底物。我们已经观察到没有任何切口的充满探针的探针显示的反切割性能要好于包含切口的形式。检查盐浓度,目标浓度和不匹配耐受性的不同实验条件,以评估探针性能。通过分别使用针对TCSV或HepBV的crRNA,针对从烟草卷曲枝病毒(TCSV)或乙型肝炎病毒(HepBV)基因组复制的dsDNA框架对Cas12a的活性进行编程。虽然按靶标活性可检测到低至10 pM dsDNA靶标,但即使在1 nM对照DNA上也未观察到离靶标活性。这项研究表明,Cas12a的反式切割不仅限于ssDNA底物,



























 京公网安备 11010802027423号
京公网安备 11010802027423号