Nature Reviews Genetics ( IF 42.7 ) Pub Date : 2020-03-31 , DOI: 10.1038/s41576-020-0224-1 Ruowang Li 1 , Yong Chen 1 , Marylyn D Ritchie 2 , Jason H Moore 1
|
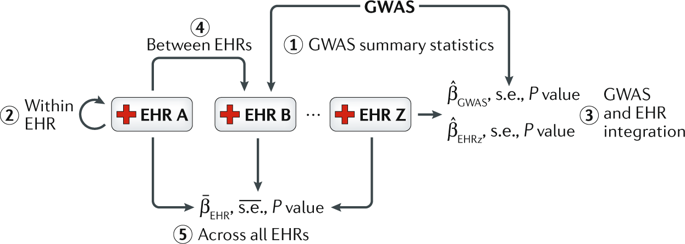
Accurate prediction of disease risk based on the genetic make-up of an individual is essential for effective prevention and personalized treatment. Nevertheless, to date, individual genetic variants from genome-wide association studies have achieved only moderate prediction of disease risk. The aggregation of genetic variants under a polygenic model shows promising improvements in prediction accuracies. Increasingly, electronic health records (EHRs) are being linked to patient genetic data in biobanks, which provides new opportunities for developing and applying polygenic risk scores in the clinic, to systematically examine and evaluate patient susceptibilities to disease. However, the heterogeneous nature of EHR data brings forth many practical challenges along every step of designing and implementing risk prediction strategies. In this Review, we present the unique considerations for using genotype and phenotype data from biobank-linked EHRs for polygenic risk prediction.
中文翻译:

用于预测疾病风险的电子健康记录和多基因风险评分。
基于个体基因构成的疾病风险准确预测对于有效预防和个性化治疗至关重要。然而,迄今为止,来自全基因组关联研究的个体遗传变异仅实现了对疾病风险的中等预测。多基因模型下遗传变异的聚集显示出预测准确性的有希望的改进。电子健康记录 (EHR) 越来越多地与生物库中的患者遗传数据相关联,这为在临床中开发和应用多基因风险评分提供了新的机会,从而系统地检查和评估患者对疾病的易感性。然而,EHR 数据的异构性质在设计和实施风险预测策略的每一步都带来了许多实际挑战。在这篇评论中,


























 京公网安备 11010802027423号
京公网安备 11010802027423号