The ISME Journal ( IF 11.0 ) Pub Date : 2020-03-27 , DOI: 10.1038/s41396-020-0635-1 Ruonan Wu 1, 2 , Benli Chai 2 , James R Cole 2, 3 , Santosh K Gunturu 2 , Xue Guo 4, 5 , Renmao Tian 4, 6 , Ji-Dong Gu 1 , Jizhong Zhou 4, 5, 7 , James M Tiedje 2, 3
|
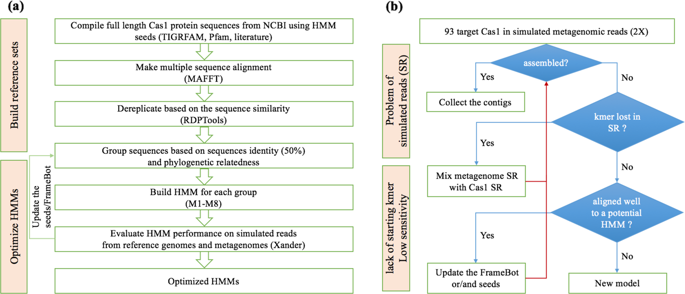
There is an increasing interest in the clustered regularly interspaced short palindromic repeats CRISPR-associated protein (CRISPR-Cas) system to reveal potential virus–host dynamics. The universal and most conserved Cas protein, cas1 is an ideal marker to elucidate CRISPR-Cas ecology. We constructed eight Hidden Markov Models (HMMs) and assembled cas1 directly from metagenomes by a targeted-gene assembler, Xander, to improve detection capacity and resolve the diverse CRISPR-Cas systems. The eight HMMs were first validated by recovering all 17 cas1 subtypes from the simulated metagenome generated from 91 prokaryotic genomes across 11 phyla. We challenged the targeted method with 48 metagenomes from a tallgrass prairie in Central Oklahoma recovering 3394 cas1. Among those, 88 were near full length, 5 times more than in de-novo assemblies from the Oklahoma metagenomes. To validate the host assignment by cas1, the targeted-assembled cas1 was mapped to the de-novo assembled contigs. All the phylum assignments of those mapped contigs were assigned independent of CRISPR-Cas genes on the same contigs and consistent with the host taxonomies predicted by the mapped cas1. We then investigated whether 8 years of soil warming altered cas1 prevalence within the communities. A shift in microbial abundances was observed during the year with the biggest temperature differential (mean 4.16 °C above ambient). cas1 prevalence increased and even in the phyla with decreased microbial abundances over the next 3 years, suggesting increasing virus–host interactions in response to soil warming. This targeted method provides an alternative means to effectively mine cas1 from metagenomes and uncover the host communities.
中文翻译:

cas1 的靶向组装表明 CRISPR-Cas 对土壤变暖的反应。
人们越来越关注集群规则间隔的短回文重复 CRISPR 相关蛋白 (CRISPR-Cas) 系统,以揭示潜在的病毒-宿主动力学。普遍且最保守的 Cas 蛋白cas1是阐明 CRISPR-Cas 生态学的理想标记。我们构建了 8 个隐马尔可夫模型 (HMM),并通过靶向基因组装器 Xander 直接从宏基因组组装cas1,以提高检测能力并解决多样化的 CRISPR-Cas 系统。首先通过从 11 个门的 91 个原核基因组生成的模拟宏基因组中恢复所有 17 个cas1亚型来验证 8 个 HMM。我们用来自俄克拉荷马州中部高草草原的 48 个宏基因组来挑战目标方法,恢复了 3394卡斯1。其中,88 个接近全长,是俄克拉荷马州宏基因组从头组装的 5 倍。为了验证 cas1 的主机分配,将目标组装的cas1映射到从头组装的contigs 。这些映射的重叠群的所有门分配都独立于相同重叠群上的 CRISPR-Cas 基因进行分配,并且与映射的cas1预测的宿主分类一致。然后,我们调查了 8 年的土壤变暖是否改变了社区内的cas1流行率。在温差最大(平均高于环境温度 4.16 °C)的一年中观察到微生物丰度的变化。cas1在接下来的 3 年中,流行率增加,甚至在微生物丰度降低的门中,这表明病毒与宿主之间的相互作用随着土壤变暖而增加。这种有针对性的方法提供了一种从宏基因组中有效挖掘cas1并发现宿主群落的替代方法。



























 京公网安备 11010802027423号
京公网安备 11010802027423号