Nature Communications ( IF 16.6 ) Pub Date : 2020-03-27 , DOI: 10.1038/s41467-020-15298-6 Ying Ma 1 , Shiquan Sun 1 , Xuequn Shang 2 , Evan T Keller 3 , Mengjie Chen 4, 5 , Xiang Zhou 1, 6
|
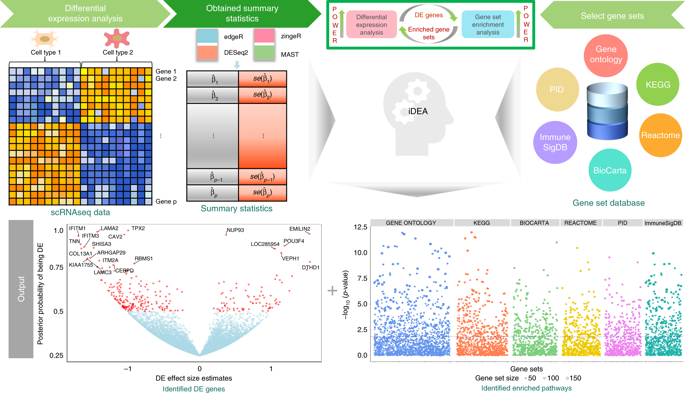
Differential expression (DE) analysis and gene set enrichment (GSE) analysis are commonly applied in single cell RNA sequencing (scRNA-seq) studies. Here, we develop an integrative and scalable computational method, iDEA, to perform joint DE and GSE analysis through a hierarchical Bayesian framework. By integrating DE and GSE analyses, iDEA can improve the power and consistency of DE analysis and the accuracy of GSE analysis. Importantly, iDEA uses only DE summary statistics as input, enabling effective data modeling through complementing and pairing with various existing DE methods. We illustrate the benefits of iDEA with extensive simulations. We also apply iDEA to analyze three scRNA-seq data sets, where iDEA achieves up to five-fold power gain over existing GSE methods and up to 64% power gain over existing DE methods. The power gain brought by iDEA allows us to identify many pathways that would not be identified by existing approaches in these data.
中文翻译:

整合差异表达和基因组富集分析,使用摘要统计数据进行scRNA-seq研究。
差异表达(DE)分析和基因集富集(GSE)分析通常用于单细胞RNA测序(scRNA-seq)研究中。在这里,我们开发了一种集成且可扩展的计算方法iDEA,以通过分层贝叶斯框架执行联合DE和GSE分析。通过整合DE和GSE分析,iDEA可以提高DE分析的功能和一致性以及GSE分析的准确性。重要的是,iDEA仅使用DE摘要统计信息作为输入,通过与各种现有DE方法进行补充和配对来实现有效的数据建模。我们通过广泛的仿真来说明iDEA的好处。我们还将iDEA应用于分析三个scRNA-seq数据集,其中iDEA的功率增益是现有GSE方法的五倍,功率增益是现有DE方法的64%。


























 京公网安备 11010802027423号
京公网安备 11010802027423号