当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
In Vitro Construction of Large-scale DNA Libraries from Fragments Containing Random Regions using Deoxyinosine-containing Oligonucleotides and Endonuclease V.
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2020-03-26 , DOI: 10.1021/acscombsci.9b00167 Yasuhide Yamamoto 1 , Takuya Terai 1 , Shigefumi Kumachi 2 , Naoto Nemoto 1, 2
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2020-03-26 , DOI: 10.1021/acscombsci.9b00167 Yasuhide Yamamoto 1 , Takuya Terai 1 , Shigefumi Kumachi 2 , Naoto Nemoto 1, 2
Affiliation
|
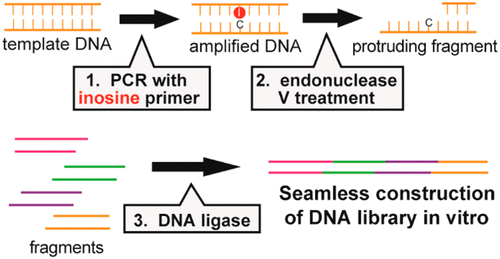
Efficient and precise construction of DNA libraries is a fundamental starting point for directed evolution of polypeptides. Recently, several in vitro selection methods have been reported that do not rely on cells for protein expression, where peptide libraries in the order of 1013 species are used for in vitro affinity selection. To maximize their potential, simple yet versatile construction of DNA libraries from several fragments containing random regions without bacterial transformation is essential. To address this issue, we herein propose a novel DNA construction methodology based on the use of polymerase chain reaction (PCR) primers containing a single deoxyinosine (I) residue near their 5' end. Treatment of the PCR products with endonuclease V generates 3' overhangs with customized lengths and sequences, which can be ligated accurately and efficiently with other fragments having exactly complementary overhangs. As a proof of concept, we constructed an artificial gene library of single-domain antibodies from four DNA fragments.
中文翻译:

使用含脱氧肌苷的寡核苷酸和核酸内切酶V从含有随机区域的片段中体外构建大规模DNA库。
高效,精确地构建DNA文库是多肽定向进化的基本起点。近来,已经报道了几种不依赖细胞表达蛋白质的体外选择方法,其中将大约1013种的肽库用于体外亲和力选择。为了最大程度地发挥其潜力,从包含细菌的随机区域的几个片段中简单而通用的DNA库构建是必不可少的。为了解决这个问题,我们在此提出一种新的DNA构建方法,该方法基于使用的聚合酶链反应(PCR)引物,在其5'端附近含有一个脱氧肌苷(I)残基。用核酸内切酶V处理PCR产物会产生3'突出端,并具有定制的长度和序列,它可以与具有完全互补的突出端的其他片段准确有效地连接。作为概念证明,我们从四个DNA片段构建了一个单域抗体的人工基因库。
更新日期:2020-04-23
中文翻译:

使用含脱氧肌苷的寡核苷酸和核酸内切酶V从含有随机区域的片段中体外构建大规模DNA库。
高效,精确地构建DNA文库是多肽定向进化的基本起点。近来,已经报道了几种不依赖细胞表达蛋白质的体外选择方法,其中将大约1013种的肽库用于体外亲和力选择。为了最大程度地发挥其潜力,从包含细菌的随机区域的几个片段中简单而通用的DNA库构建是必不可少的。为了解决这个问题,我们在此提出一种新的DNA构建方法,该方法基于使用的聚合酶链反应(PCR)引物,在其5'端附近含有一个脱氧肌苷(I)残基。用核酸内切酶V处理PCR产物会产生3'突出端,并具有定制的长度和序列,它可以与具有完全互补的突出端的其他片段准确有效地连接。作为概念证明,我们从四个DNA片段构建了一个单域抗体的人工基因库。

























 京公网安备 11010802027423号
京公网安备 11010802027423号