Cell Systems ( IF 9.3 ) Pub Date : 2020-03-25 , DOI: 10.1016/j.cels.2020.02.009 Peter Orchard 1 , Yasuhiro Kyono 2 , John Hensley 1 , Jacob O Kitzman 2 , Stephen C J Parker 2
|
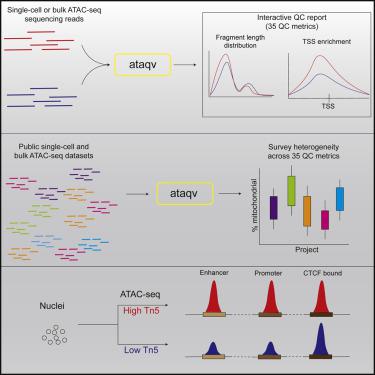
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) has become the preferred method for mapping chromatin accessibility due to its time and input material efficiency. However, it can be difficult to evaluate data quality and identify sources of technical bias across samples. Here, we present ataqv, a computational toolkit for efficiently measuring, visualizing, and comparing quality control (QC) results across samples and experiments. We use ataqv to analyze 2,009 public ATAC-seq datasets; their QC metrics display a 10-fold range. Tn5 dosage experiments and statistical modeling show that technical variation in the ratio of Tn5 transposase to nuclei and sequencing flowcell density induces systematic bias in ATAC-seq data by changing the enrichment of reads across functional genomic annotations including promoters, enhancers, and transcription-factor-bound regions, with the notable exception of CTCF. ataqv can be integrated into existing computational pipelines and is freely available at https://github.com/ParkerLab/ataqv/.
中文翻译:

带有ataqv的ATAC-Seq数据中偏差的量化,动态可视化和验证。
由于测序时间和输入材料的效率,使用测序法(ATAC-seq)进行转座酶可及性染色质的测定已成为映射染色质可及性的首选方法。但是,可能难以评估数据质量并确定样本间技术偏见的来源。在这里,我们介绍ataqv,这是一种用于有效测量,可视化和比较样品和实验中质量控制(QC)结果的计算工具包。我们使用ataqv分析2,009个公共ATAC-seq数据集;他们的质量控制指标显示10倍范围。Tn5剂量实验和统计模型表明,Tn5转座酶与细胞核比例和流通池密度的技术变化可通过改变功能基因组注释(包括启动子,增强子,和转录因子结合区,CTCF除外。ataqv可以集成到现有的计算管道中,并且可以从https://github.com/ParkerLab/ataqv/免费获得。

























 京公网安备 11010802027423号
京公网安备 11010802027423号