当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Accurately Predicting Mutation-Caused Stability Changes from Protein Sequences Using Extreme Gradient Boosting.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jcim.0c00064 Xuan Lv 1 , Jianwen Chen 2 , Yutong Lu 2 , Zhiguang Chen 2 , Nong Xiao 1, 2 , Yuedong Yang 2, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jcim.0c00064 Xuan Lv 1 , Jianwen Chen 2 , Yutong Lu 2 , Zhiguang Chen 2 , Nong Xiao 1, 2 , Yuedong Yang 2, 3
Affiliation
|
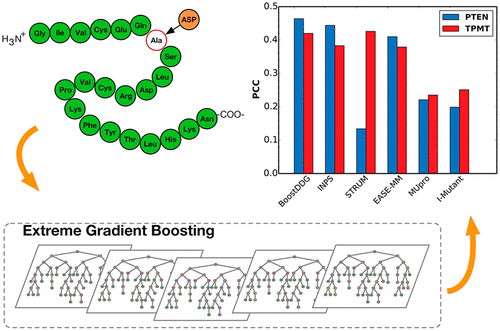
Accurately predicting the impact of point mutation on protein stability has crucial roles in protein design and engineering. In this study, we proposed a novel method (BoostDDG) to predict stability changes upon point mutations from protein sequences based on the extreme gradient boosting. We extracted features comprehensively from evolutional information and predicted structures and performed feature selection by a strategy of sequential forward selection. The features and parameters were optimized by homologue-based cross-validation to avoid overfitting. Finally, we found that 14 features from six groups led to the highest Pearson correlation coefficient (PCC) of 0.535, which is consistent with the 0.540 on an independent test. Our method was indicated to consistently outperform other sequence-based methods on three precompiled test sets, and 7363 variants on two proteins (PTEN and TPMT). These results highlighted that BoostDDG is a powerful tool for predicting stability changes upon point mutations from protein sequences.
中文翻译:

使用极端梯度增强从蛋白质序列准确预测突变引起的稳定性变化。
准确预测点突变对蛋白质稳定性的影响在蛋白质设计和工程中至关重要。在这项研究中,我们提出了一种新方法(BoostDDG),可基于极端梯度增强预测蛋白质序列中点突变后的稳定性变化。我们从进化信息和预测结构中全面提取了特征,并通过顺序正向选择策略进行了特征选择。通过基于同源物的交叉验证对特征和参数进行了优化,以避免过度拟合。最后,我们发现六组中的14个特征导致最高的Pearson相关系数(PCC)为0.535,这与独立测试中的0.540一致。我们的方法在三个预编译的测试集上始终优于其他基于序列的方法,和两种蛋白质(PTEN和TPMT)上的7363变体。这些结果表明,BoostDDG是预测蛋白质序列中点突变后稳定性变化的强大工具。
更新日期:2020-03-23
中文翻译:

使用极端梯度增强从蛋白质序列准确预测突变引起的稳定性变化。
准确预测点突变对蛋白质稳定性的影响在蛋白质设计和工程中至关重要。在这项研究中,我们提出了一种新方法(BoostDDG),可基于极端梯度增强预测蛋白质序列中点突变后的稳定性变化。我们从进化信息和预测结构中全面提取了特征,并通过顺序正向选择策略进行了特征选择。通过基于同源物的交叉验证对特征和参数进行了优化,以避免过度拟合。最后,我们发现六组中的14个特征导致最高的Pearson相关系数(PCC)为0.535,这与独立测试中的0.540一致。我们的方法在三个预编译的测试集上始终优于其他基于序列的方法,和两种蛋白质(PTEN和TPMT)上的7363变体。这些结果表明,BoostDDG是预测蛋白质序列中点突变后稳定性变化的强大工具。



























 京公网安备 11010802027423号
京公网安备 11010802027423号