PLOS Biology ( IF 9.8 ) Pub Date : 2020-03-20 , DOI: 10.1371/journal.pbio.3000661 Travis J Wiles 1 , Brandon H Schlomann 1, 2 , Elena S Wall 1 , Reina Betancourt 1 , Raghuveer Parthasarathy 1, 2 , Karen Guillemin 1, 3
|
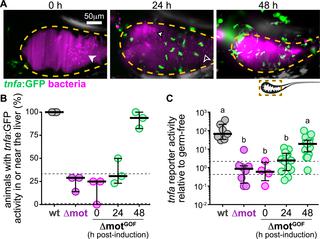
Some of the densest microbial ecosystems in nature thrive within the intestines of humans and other animals. To protect mucosal tissues and maintain immune tolerance, animal hosts actively sequester bacteria within the intestinal lumen. In response, numerous bacterial pathogens and pathobionts have evolved strategies to subvert spatial restrictions, thereby undermining immune homeostasis. However, in many cases, it is unclear how escaping host spatial control benefits gut bacteria and how changes in intestinal biogeography are connected to inflammation. A better understanding of these processes could uncover new targets for treating microbiome-mediated inflammatory diseases. To this end, we investigated the spatial organization and dynamics of bacterial populations within the intestine using larval zebrafish and live imaging. We discovered that a proinflammatory Vibrio symbiont native to zebrafish governs its own spatial organization using swimming motility and chemotaxis. Surprisingly, we found that Vibrio’s motile behavior does not enhance its growth rate but rather promotes its persistence by enabling it to counter intestinal flow. In contrast, Vibrio mutants lacking motility traits surrender to host spatial control, becoming aggregated and entrapped within the lumen. Consequently, nonmotile and nonchemotactic mutants are susceptible to intestinal expulsion and experience large fluctuations in absolute abundance. Further, we found that motile Vibrio cells induce expression of the proinflammatory cytokine tumor necrosis factor alpha (TNFα) in gut-associated macrophages and the liver. Using inducible genetic switches, we demonstrate that swimming motility can be manipulated in situ to modulate the spatial organization, persistence, and inflammatory activity of gut bacterial populations. Together, our findings suggest that host spatial control over resident microbiota plays a broader role in regulating the abundance and persistence of gut bacteria than simply protecting mucosal tissues. Moreover, we show that intestinal flow and bacterial motility are potential targets for therapeutically managing bacterial spatial organization and inflammatory activity within the gut.
中文翻译:

肠道细菌共生体的游泳运动可增强对肠道驱逐的抵抗力,并增强炎症。
自然界中一些最稠密的微生物生态系统在人类和其他动物的肠道内壮成长。为了保护粘膜组织并保持免疫耐受性,动物宿主主动在肠道腔内隔离细菌。作为响应,许多细菌病原体和病原生物已经进化出了破坏空间限制从而破坏免疫稳态的策略。然而,在许多情况下,尚不清楚逃避宿主空间控制如何使肠道细菌受益,以及肠道生物地理学的变化如何与炎症相关。对这些过程的更好的了解可能会发现治疗微生物组介导的炎性疾病的新目标。为此,我们使用幼虫斑马鱼和实时成像技术研究了肠道内细菌种群的空间组织和动态。斑马鱼固有的共生弧菌利用游泳运动和趋化性来控制其自身的空间组织。令人惊讶的是,我们发现弧菌的运动行为不会提高其生长速度,而是通过使其能够抵抗肠道流动来促进其持久性。相反,缺乏运动性状的弧菌突变体向宿主空间控制投降,在腔内聚集并被捕获。因此,非运动型和非趋化性突变体容易被肠道驱逐,并且绝对丰度发生较大波动。此外,我们发现运动型弧菌细胞诱导肠道相关巨噬细胞和肝脏中促炎性细胞因子肿瘤坏死因子α(TNFα)的表达。使用诱导型遗传开关,我们证明了游泳运动可以被原位操纵,以调节肠道细菌种群的空间组织,持久性和炎症活性。总之,我们的发现表明,宿主对驻留微生物群的空间控制在调节肠道细菌的丰度和持久性方面起着比单纯保护粘膜组织更广泛的作用。此外,我们表明肠流量和细菌运动性是治疗性管理肠道内细菌空间组织和炎症活动的潜在目标。


























 京公网安备 11010802027423号
京公网安备 11010802027423号