当前位置:
X-MOL 学术
›
Syst. Entomol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The phylogeny of insects in the data-driven era
Systematic Entomology ( IF 4.8 ) Pub Date : 2019-12-19 , DOI: 10.1111/syen.12414 Douglas Chesters 1
Systematic Entomology ( IF 4.8 ) Pub Date : 2019-12-19 , DOI: 10.1111/syen.12414 Douglas Chesters 1
Affiliation
|
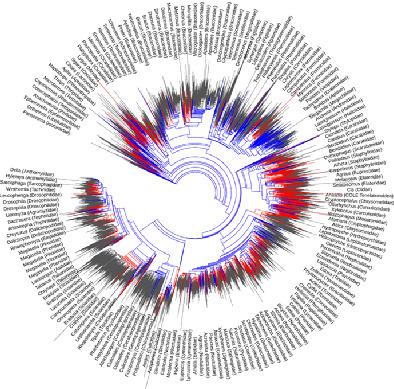
Maturation of omics and DNA barcode programs along with advances in sequence analysis tools and phyloinformatics protocols are enabling the realization of comprehensive and robust phylogenies of even the most diverse lineages. Several lineages in insects have undergone hyper‐radiations, and thus a unified picture of their evolution is ultimately required for understanding the process of diversification. In this study I further develop informatics protocols for de novo phylogenetic construction, and present the most species‐comprehensive insect phylogeny to date, constructed hierarchically using c. 440 transcriptomes, 1490 mitogenomes, DNA barcodes for 69 000 species, and several additional species‐rich markers. Even with this expanded transcriptome backbone, support is still insufficient for some historically problematic nodes, particularly in Polyneoptera and Paraneoptera. Low support (measured by internode certainty) was observed for the node separating Dictyoptera from its sister polyneopeterans; configuration of the Paraneoptera was not resolved; the recently proposed Hymenoptera grouping Eusymphyta received high support, while Parasitoida did not; and Orthorrhapha (Diptera) was not recovered. Sampling is uneven across the insects, and while highly sequenced lineages (e.g. Lepidoptera) boast greater information content, this accompanies computational burdens. The protocol and resulting tree represent an advance in the analytic and phylogenetic framework, for an objectively and consistently determined species‐comprehensive phylogeny.
中文翻译:

数据驱动时代昆虫的系统发育
组学和 DNA 条形码程序的成熟,以及序列分析工具和系统信息学协议的进步,使得即使是最多样化的谱系也能实现全面而强大的系统发育。昆虫中的几个谱系经历了超辐射,因此最终需要对它们的进化有统一的了解,以了解多样化的过程。在这项研究中,我进一步开发了用于从头系统发育构建的信息学协议,并展示了迄今为止最全面的昆虫系统发育,使用 c. 分层构建。440 个转录组、1490 个有丝分裂基因组、69 000 个物种的 DNA 条形码,以及几个额外的物种丰富的标记。即使有了这个扩展的转录组主干,对一些历史上有问题的节点的支持仍然不足,特别是在多翅目和副翅目中。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度测序的谱系(例如鳞翅目)拥有更多的信息内容,但这伴随着计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。
更新日期:2019-12-19
中文翻译:

数据驱动时代昆虫的系统发育
组学和 DNA 条形码程序的成熟,以及序列分析工具和系统信息学协议的进步,使得即使是最多样化的谱系也能实现全面而强大的系统发育。昆虫中的几个谱系经历了超辐射,因此最终需要对它们的进化有统一的了解,以了解多样化的过程。在这项研究中,我进一步开发了用于从头系统发育构建的信息学协议,并展示了迄今为止最全面的昆虫系统发育,使用 c. 分层构建。440 个转录组、1490 个有丝分裂基因组、69 000 个物种的 DNA 条形码,以及几个额外的物种丰富的标记。即使有了这个扩展的转录组主干,对一些历史上有问题的节点的支持仍然不足,特别是在多翅目和副翅目中。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度测序的谱系(例如鳞翅目)拥有更多的信息内容,但这伴随着计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。观察到将网翅目与其姊妹多翅目分开的节点的支持度低(通过节间确定性衡量);副翅目的配置没有解决;最近提出的膜翅目属群 Eusymphyta 得到了很高的支持,而 Parasitoida 则没有;和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。和 Orthorrhapha(双翅目)没有被回收。昆虫的采样是不均匀的,虽然高度排序的谱系(例如鳞翅目)拥有更多的信息内容,但这会带来计算负担。该协议和生成的树代表了分析和系统发育框架的进步,用于客观和一致确定的物种综合系统发育。



























 京公网安备 11010802027423号
京公网安备 11010802027423号