当前位置:
X-MOL 学术
›
J. Mol. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Bioinformatic Analysis of the Flavin-Dependent Amine Oxidase Superfamily: Adaptations for Substrate Specificity and Catalytic Diversity.
Journal of Molecular Biology ( IF 5.6 ) Pub Date : 2020-03-19 , DOI: 10.1016/j.jmb.2020.03.007 Margarita A Tararina 1 , Karen N Allen 2
Journal of Molecular Biology ( IF 5.6 ) Pub Date : 2020-03-19 , DOI: 10.1016/j.jmb.2020.03.007 Margarita A Tararina 1 , Karen N Allen 2
Affiliation
|
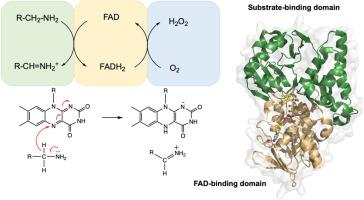
The flavin-dependent amine oxidase (FAO) superfamily consists of over 9000 nonredundant sequences represented in all domains of life. Of the thousands of members identified, only 214 have been functionally annotated to date, and 40 unique structures are represented in the Protein Data Bank. The few functionally characterized members share a catalytic mechanism involving the oxidation of an amine substrate through transfer of a hydride to the FAD cofactor, with differences observed in substrate specificities. Previous studies have focused on comparing a subset of superfamily members. Here, we present a comprehensive analysis of the FAO superfamily based on reaction mechanism and substrate recognition. Using a dataset of 9192 sequences, a sequence similarity network, and subsequently, a genome neighborhood network were constructed, organizing the superfamily into eight subgroups that accord with substrate type. Likewise, through phylogenetic analysis, the evolutionary relationship of subgroups was determined, delineating the divergence between enzymes based on organism, substrate, and mechanism. In addition, using sequences and atomic coordinates of 22 structures from the Protein Data Bank to perform sequence and structural alignments, active-site elements were identified, showing divergence from the canonical aromatic-cage residues to accommodate large substrates. These specificity determinants are held in a structural framework comprising a core domain catalyzing the oxidation of amines with an auxiliary domain for substrate recognition. Overall, analysis of the FAO superfamily reveals a modular fold with cofactor and substrate-binding domains allowing for diversity of recognition via insertion/deletions. This flexibility allows facile evolution of new activities, as shown by reinvention of function between subfamilies.
中文翻译:

黄素依赖性胺氧化酶超家族的生物信息学分析:底物特异性和催化多样性的适应。
黄素依赖性胺氧化酶(FAO)超家族由生活中所有域中代表的9000多个非冗余序列组成。迄今为止,在鉴定出的数千个成员中,只有214个在功能上得到了注释,蛋白质数据库中代表了40个独特的结构。少数具有功能特征的成员共享催化机制,该机制涉及通过将氢化物转移至FAD辅因子来氧化胺底物,同时观察到底物特异性的差异。先前的研究集中于比较超家族成员的子集。在这里,我们根据反应机理和底物识别对粮农组织超家族进行全面分析。使用9192个序列的数据集,构建了一个序列相似性网络,然后构建了一个基因组邻域网络,将超家族分为与底物类型相符的八个亚组。同样,通过系统发育分析,确定了亚组的进化关系,根据生物,底物和机制描绘了酶之间的差异。此外,使用蛋白质数据库中22种结构的序列和原子坐标进行序列和结构比对,鉴定了活性位点元素,显示出与规范的芳族笼残基不同,可容纳较大的底物。这些特异性决定子被保持在包含骨架结构域的结构框架中,该核心结构域催化具有用于底物识别的辅助结构域的胺的氧化。总体,对粮农组织超家族的分析显示,辅因子和底物结合域具有模块折叠性,可通过插入/缺失实现识别多样性。这种灵活性允许新活动的轻松演变,如子家族之间功能的重新发明所示。
更新日期:2020-03-19
中文翻译:

黄素依赖性胺氧化酶超家族的生物信息学分析:底物特异性和催化多样性的适应。
黄素依赖性胺氧化酶(FAO)超家族由生活中所有域中代表的9000多个非冗余序列组成。迄今为止,在鉴定出的数千个成员中,只有214个在功能上得到了注释,蛋白质数据库中代表了40个独特的结构。少数具有功能特征的成员共享催化机制,该机制涉及通过将氢化物转移至FAD辅因子来氧化胺底物,同时观察到底物特异性的差异。先前的研究集中于比较超家族成员的子集。在这里,我们根据反应机理和底物识别对粮农组织超家族进行全面分析。使用9192个序列的数据集,构建了一个序列相似性网络,然后构建了一个基因组邻域网络,将超家族分为与底物类型相符的八个亚组。同样,通过系统发育分析,确定了亚组的进化关系,根据生物,底物和机制描绘了酶之间的差异。此外,使用蛋白质数据库中22种结构的序列和原子坐标进行序列和结构比对,鉴定了活性位点元素,显示出与规范的芳族笼残基不同,可容纳较大的底物。这些特异性决定子被保持在包含骨架结构域的结构框架中,该核心结构域催化具有用于底物识别的辅助结构域的胺的氧化。总体,对粮农组织超家族的分析显示,辅因子和底物结合域具有模块折叠性,可通过插入/缺失实现识别多样性。这种灵活性允许新活动的轻松演变,如子家族之间功能的重新发明所示。


























 京公网安备 11010802027423号
京公网安备 11010802027423号