iScience ( IF 5.8 ) Pub Date : 2020-03-19 , DOI: 10.1016/j.isci.2020.100991 Hao Lv 1 , Fu-Ying Dao 1 , Dan Zhang 1 , Zheng-Xing Guan 1 , Hui Yang 1 , Wei Su 1 , Meng-Lu Liu 1 , Hui Ding 1 , Wei Chen 2 , Hao Lin 1
|
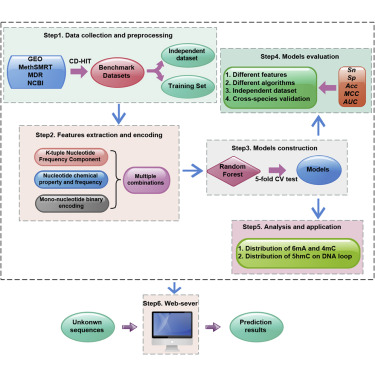
5hmC, 6mA, and 4mC are three common DNA modifications and are involved in various of biological processes. Accurate genome-wide identification of these sites is invaluable for better understanding their biological functions. Owing to the labor-intensive and expensive nature of experimental methods, it is urgent to develop computational methods for the genome-wide detection of these sites. Keeping this in mind, the current study was devoted to construct a computational method to identify 5hmC, 6mA, and 4mC. We initially used K-tuple nucleotide component, nucleotide chemical property and nucleotide frequency, and mono-nucleotide binary encoding scheme to formulate samples. Subsequently, random forest was utilized to identify 5hmC, 6mA, and 4mC sites. Cross-validated results showed that the proposed method could produce the excellent generalization ability in the identification of the three modification sites. Based on the proposed model, a web-server called iDNA-MS was established and is freely accessible at http://lin-group.cn/server/iDNA-MS.
中文翻译:

iDNA-MS:用于检测多个基因组中DNA修饰位点的集成计算工具。
5hmC,6mA和4mC是三种常见的DNA修饰,涉及各种生物过程。这些位点的全基因组准确鉴定对于更好地了解其生物学功能非常重要。由于实验方法的劳动强度大和昂贵的性质,迫切需要开发用于在全基因组范围内检测这些位点的计算方法。牢记这一点,当前的研究致力于构建一种识别5hmC,6mA和4mC的计算方法。我们最初使用K-元组核苷酸成分,核苷酸化学性质和核苷酸频率,以及单核苷酸二进制编码方案来配制样品。随后,利用随机森林来识别5hmC,6mA和4mC的位点。交叉验证的结果表明,该方法在三个修饰位点的识别中具有出色的泛化能力。基于提出的模型,建立了一个名为iDNA-MS的网络服务器,可以从http://lin-group.cn/server/iDNA-MS免费访问。


























 京公网安备 11010802027423号
京公网安备 11010802027423号