Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Highly Parallel Profiling of Cas9 Variant Specificity.
Molecular Cell ( IF 16.0 ) Pub Date : 2020-03-17 , DOI: 10.1016/j.molcel.2020.02.023 Jonathan L Schmid-Burgk 1 , Linyi Gao 1 , David Li 1 , Zachary Gardner 1 , Jonathan Strecker 1 , Blake Lash 1 , Feng Zhang 2
Molecular Cell ( IF 16.0 ) Pub Date : 2020-03-17 , DOI: 10.1016/j.molcel.2020.02.023 Jonathan L Schmid-Burgk 1 , Linyi Gao 1 , David Li 1 , Zachary Gardner 1 , Jonathan Strecker 1 , Blake Lash 1 , Feng Zhang 2
Affiliation
|
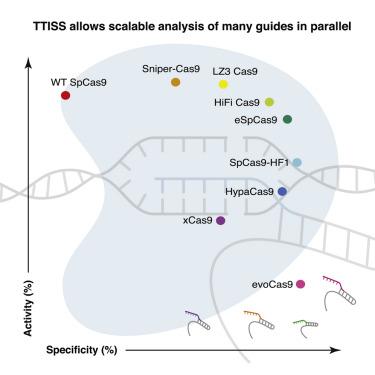
Determining the off-target cleavage profile of programmable nucleases is an important consideration for any genome editing experiment, and a number of Cas9 variants have been reported that improve specificity. We describe here tagmentation-based tag integration site sequencing (TTISS), an efficient, scalable method for analyzing double-strand breaks (DSBs) that we apply in parallel to eight Cas9 variants across 59 targets. Additionally, we generated thousands of other Cas9 variants and screened for variants with enhanced specificity and activity, identifying LZ3 Cas9, a high specificity variant with a unique +1 insertion profile. This comprehensive comparison reveals a general trade-off between Cas9 activity and specificity and provides information about the frequency of generation of +1 insertions, which has implications for correcting frameshift mutations.
中文翻译:

Cas9变异特异性的高度并行分析。
确定可编程核酸酶的脱靶裂解谱是任何基因组编辑实验的重要考虑因素,并且已经报道了许多可提高特异性的Cas9变体。我们在这里描述基于标签的标签整合位点测序(TTISS),这是一种有效的,可扩展的分析双链断裂(DSB)的方法,我们将其并行应用于跨59个靶标的8个Cas9变体。此外,我们生成了数千个其他Cas9变体,并筛选了具有增强的特异性和活性的变体,从而确定了LZ3 Cas9,这是一种具有独特+1插入模式的高特异性变体。这项全面的比较揭示了Cas9活性和特异性之间的一般平衡,并提供了有关+1插入产生频率的信息,
更新日期:2020-03-17
中文翻译:

Cas9变异特异性的高度并行分析。
确定可编程核酸酶的脱靶裂解谱是任何基因组编辑实验的重要考虑因素,并且已经报道了许多可提高特异性的Cas9变体。我们在这里描述基于标签的标签整合位点测序(TTISS),这是一种有效的,可扩展的分析双链断裂(DSB)的方法,我们将其并行应用于跨59个靶标的8个Cas9变体。此外,我们生成了数千个其他Cas9变体,并筛选了具有增强的特异性和活性的变体,从而确定了LZ3 Cas9,这是一种具有独特+1插入模式的高特异性变体。这项全面的比较揭示了Cas9活性和特异性之间的一般平衡,并提供了有关+1插入产生频率的信息,



























 京公网安备 11010802027423号
京公网安备 11010802027423号