当前位置:
X-MOL 学术
›
Cell Host Microbe
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2.
Cell Host & Microbe ( IF 30.3 ) Pub Date : 2020-03-16 , DOI: 10.1016/j.chom.2020.03.002 Alba Grifoni 1 , John Sidney 1 , Yun Zhang 2 , Richard H Scheuermann 3 , Bjoern Peters 4 , Alessandro Sette 4
Cell Host & Microbe ( IF 30.3 ) Pub Date : 2020-03-16 , DOI: 10.1016/j.chom.2020.03.002 Alba Grifoni 1 , John Sidney 1 , Yun Zhang 2 , Richard H Scheuermann 3 , Bjoern Peters 4 , Alessandro Sette 4
Affiliation
|
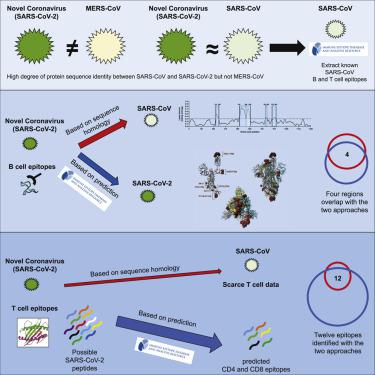
Effective countermeasures against the recent emergence and rapid expansion of the 2019 novel coronavirus (SARS-CoV-2) require the development of data and tools to understand and monitor its spread and immune responses to it. However, little information is available about the targets of immune responses to SARS-CoV-2. We used the Immune Epitope Database and Analysis Resource (IEDB) to catalog available data related to other coronaviruses. This includes SARS-CoV, which has high sequence similarity to SARS-CoV-2 and is the best-characterized coronavirus in terms of epitope responses. We identified multiple specific regions in SARS-CoV-2 that have high homology to the SARS-CoV virus. Parallel bioinformatic predictions identified a priori potential B and T cell epitopes for SARS-CoV-2. The independent identification of the same regions using two approaches reflects the high probability that these regions are promising targets for immune recognition of SARS-CoV-2. These predictions can facilitate effective vaccine design against this virus of high priority.
中文翻译:

序列同源性和生物信息学方法可以预测对 SARS-CoV-2 的免疫反应的候选目标。
针对 2019 年新型冠状病毒 (SARS-CoV-2) 最近出现和迅速扩散的有效对策需要开发数据和工具来了解和监测其传播和免疫反应。然而,关于对 SARS-CoV-2 的免疫反应目标的信息很少。我们使用免疫表位数据库和分析资源 (IEDB) 对与其他冠状病毒相关的可用数据进行分类。这包括 SARS-CoV,它与 SARS-CoV-2 具有高度序列相似性,是表位反应方面特征最好的冠状病毒。我们在 SARS-CoV-2 中发现了多个与 SARS-CoV 病毒具有高度同源性的特定区域。平行生物信息学预测确定了 SARS-CoV-2 的先验潜在 B 和 T 细胞表位。使用两种方法独立识别相同区域反映了这些区域很有可能成为 SARS-CoV-2 免疫识别的有希望的目标。这些预测可以促进针对这种高优先级病毒的有效疫苗设计。
更新日期:2020-04-20
中文翻译:

序列同源性和生物信息学方法可以预测对 SARS-CoV-2 的免疫反应的候选目标。
针对 2019 年新型冠状病毒 (SARS-CoV-2) 最近出现和迅速扩散的有效对策需要开发数据和工具来了解和监测其传播和免疫反应。然而,关于对 SARS-CoV-2 的免疫反应目标的信息很少。我们使用免疫表位数据库和分析资源 (IEDB) 对与其他冠状病毒相关的可用数据进行分类。这包括 SARS-CoV,它与 SARS-CoV-2 具有高度序列相似性,是表位反应方面特征最好的冠状病毒。我们在 SARS-CoV-2 中发现了多个与 SARS-CoV 病毒具有高度同源性的特定区域。平行生物信息学预测确定了 SARS-CoV-2 的先验潜在 B 和 T 细胞表位。使用两种方法独立识别相同区域反映了这些区域很有可能成为 SARS-CoV-2 免疫识别的有希望的目标。这些预测可以促进针对这种高优先级病毒的有效疫苗设计。


























 京公网安备 11010802027423号
京公网安备 11010802027423号