当前位置:
X-MOL 学术
›
Comp. Biochem. Physiol. C Toxicol. Pharmacol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Interspecific and intraspecific venom enzymatic variation among cobras (Naja sp. and Ophiophagus hannah).
Comparative Biochemistry and Physiology C: Toxicology & Pharmacology ( IF 3.9 ) Pub Date : 2020-03-16 , DOI: 10.1016/j.cbpc.2020.108743 Cassandra M Modahl 1 , Amir Roointan 2 , Jessica Rogers 3 , Katelyn Currier 3 , Stephen P Mackessy 3
Comparative Biochemistry and Physiology C: Toxicology & Pharmacology ( IF 3.9 ) Pub Date : 2020-03-16 , DOI: 10.1016/j.cbpc.2020.108743 Cassandra M Modahl 1 , Amir Roointan 2 , Jessica Rogers 3 , Katelyn Currier 3 , Stephen P Mackessy 3
Affiliation
|
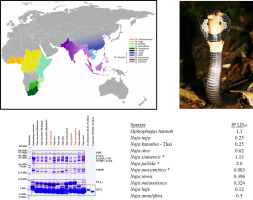
The genera Ophiophagus and Naja comprise part of a clade of snakes referred to as cobras, dangerously venomous front-fanged snakes in the family Elapidae responsible for significant human mortality and morbidity throughout Asia and Africa. We evaluated venom enzyme variation for eleven cobra species and three N. kaouthia populations using SDS-PAGE venom fingerprinting and numerous enzyme assays. Acetylcholinesterase and PLA2 activities were the most variable between species, and PLA2 activity was significantly different between Malaysian and Thailand N. kaouthia populations. Venom metalloproteinase activity was low and significantly different among most species, but levels were identical for N. kaouthia populations; minor variation in venom L-amino acid oxidase and phosphodiesterase activities were seen between cobra species. Naja siamensis venom lacked the α-fibrinogenolytic activity common to other cobra venoms. In addition, venom from N. siamensis had no detectable metalloproteinase activity and exhibited an SDS-PAGE profile with reduced abundance of higher mass proteins. Venom profiles from spitting cobras (N. siamensis, N. pallida, and N. mossambica) exhibited similar reductions in higher mass proteins, suggesting the evolution of venoms of reduced complexity and decreased enzymatic activity among spitting cobras. Generally, the venom proteomes of cobras show highly abundant three-finger toxin diversity, followed by large quantities of PLA2s. However, PLA2 bands and activity were very reduced for N. haje, N. annulifera and N. nivea. Venom compositionalenzy analysis provides insight into the evolution, diversification and distribution of different venom phenotypes that complements venomic data, and this information is critical for the development of effective antivenoms and snakebite treatment.
中文翻译:

眼镜蛇之间的种间和种内毒液酶促变化(Naja sp。和Ophiophagus hannah)。
眼镜蛇属和眼镜蛇属是蛇类的一部分,被称为眼镜蛇,是蛇蝎科中毒蛇毒的毒蛇,负责整个亚洲和非洲的人类死亡和发病。我们使用SDS-PAGE毒液指纹图谱和大量酶分析方法,评估了11种眼镜蛇物种和3个猪笼草N. kaouthia种群的毒液酶变化。乙酰胆碱酯酶和PLA2活性在物种之间变化最大,而马来西亚和泰国南北香茅种群之间的PLA2活性显着不同。毒液金属蛋白酶活性低,在大多数物种中差异显着,但对于长叶猪笼草种群而言,其水平是相同的。眼镜蛇物种之间的毒液L-氨基酸氧化酶和磷酸二酯酶活性的微小变化。暹罗眼镜蛇毒缺乏其他眼镜蛇毒常见的α-纤维蛋白原分解活性。此外,来自暹罗猪笼草的毒液没有可检测到的金属蛋白酶活性,并且表现出SDS-PAGE图谱,而大量高质量的蛋白质却减少了。吐痰眼镜蛇(N. siamensis,N。pallida和N. mossambica)的毒液谱显示出类似的高质量蛋白质减少,表明吐痰的眼镜蛇中复杂性降低和酶活性降低的毒液进化。通常,眼镜蛇的蛇毒蛋白质组显示出高度丰富的三指毒素多样性,其次是大量的PLA2。但是,哈氏猪笼草,环孢猪笼草和妮维猪笼草的PLA2条带和活性均大大降低。毒液成分酶分析可深入了解进化过程,
更新日期:2020-03-16
中文翻译:

眼镜蛇之间的种间和种内毒液酶促变化(Naja sp。和Ophiophagus hannah)。
眼镜蛇属和眼镜蛇属是蛇类的一部分,被称为眼镜蛇,是蛇蝎科中毒蛇毒的毒蛇,负责整个亚洲和非洲的人类死亡和发病。我们使用SDS-PAGE毒液指纹图谱和大量酶分析方法,评估了11种眼镜蛇物种和3个猪笼草N. kaouthia种群的毒液酶变化。乙酰胆碱酯酶和PLA2活性在物种之间变化最大,而马来西亚和泰国南北香茅种群之间的PLA2活性显着不同。毒液金属蛋白酶活性低,在大多数物种中差异显着,但对于长叶猪笼草种群而言,其水平是相同的。眼镜蛇物种之间的毒液L-氨基酸氧化酶和磷酸二酯酶活性的微小变化。暹罗眼镜蛇毒缺乏其他眼镜蛇毒常见的α-纤维蛋白原分解活性。此外,来自暹罗猪笼草的毒液没有可检测到的金属蛋白酶活性,并且表现出SDS-PAGE图谱,而大量高质量的蛋白质却减少了。吐痰眼镜蛇(N. siamensis,N。pallida和N. mossambica)的毒液谱显示出类似的高质量蛋白质减少,表明吐痰的眼镜蛇中复杂性降低和酶活性降低的毒液进化。通常,眼镜蛇的蛇毒蛋白质组显示出高度丰富的三指毒素多样性,其次是大量的PLA2。但是,哈氏猪笼草,环孢猪笼草和妮维猪笼草的PLA2条带和活性均大大降低。毒液成分酶分析可深入了解进化过程,



























 京公网安备 11010802027423号
京公网安备 11010802027423号