当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Alteration of Transcriptional Regulator Rob In Vivo: Enhancement of Promoter DNA Binding and Antibiotic Resistance in the Presence of Nucleobase Amino Acids.
Biochemistry ( IF 2.9 ) Pub Date : 2020-03-17 , DOI: 10.1021/acs.biochem.0c00103 Chao Zhang 1 , Shengxi Chen 1 , Xiaoguang Bai 1 , Larisa M Dedkova 1 , Sidney M Hecht 1
Biochemistry ( IF 2.9 ) Pub Date : 2020-03-17 , DOI: 10.1021/acs.biochem.0c00103 Chao Zhang 1 , Shengxi Chen 1 , Xiaoguang Bai 1 , Larisa M Dedkova 1 , Sidney M Hecht 1
Affiliation
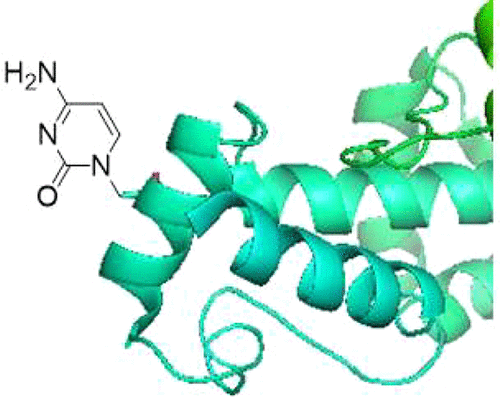
|
The identification of proteins that bind selectively to nucleic acid sequences is an ongoing challenge. We previously synthesized nucleobase amino acids designed to replace proteinogenic amino acids; these were incorporated into proteins to bind specific nucleic acids predictably. An early example involved selective cell free binding of the hnRNP LL RRM1 domain to its i-motif DNA target via Watson-Crick-like H-bonding interactions. In this study, we employ the X-ray crystal structure of transcriptional regulator Rob bound to its micF promoter, which occurred without DNA distortion. Rob proteins modified in vivo with nucleobase amino acids at position 40 exhibited altered DNA promoter binding, as predicted on the basis of their Watson-Crick-like H-bonding interactions with promoter DNA A-box residue Gua-6. Rob protein expression ultimately controls phenotypic changes, including resistance to antibiotics. Although Rob proteins with nucleobase amino acids were expressed in Escherichia coli at levels estimated to be only a fraction of that of the wild-type Rob protein, those modified proteins that bound to the micF promoter more avidly than the wild type in vitro also produced greater resistance to macrolide antibiotics roxithromycin and clarithromycin in vivo, as well as the β-lactam antibiotic ampicillin. Also demonstrated is the statistical significance of altered DNA binding and antibiotic resistance for key Rob analogues. These preliminary findings suggest the ultimate utility of nucleobase amino acids in altering and controlling preferred nucleic acid target sequences by proteins, for probing molecular interactions critical to protein function, and for enhancing phenotypic changes in vivo by regulatory protein analogues.
中文翻译:

体内转录调节剂Rob的改变:在存在核碱基氨基酸的情况下增强启动子DNA结合和抗生素抗性。
选择性结合核酸序列的蛋白质的鉴定是一个持续的挑战。我们先前合成了旨在取代蛋白原性氨基酸的核碱基氨基酸;将它们掺入蛋白质以可预测地结合特异性核酸。一个较早的例子涉及hnRNP LL RRM1结构域通过Watson-Crick样的H键相互作用选择性地无细胞结合其i-基序DNA靶标。在这项研究中,我们采用了转录调节器Rob的X射线晶体结构,该结构与其micF启动子结合,而DNA不会发生畸变。如在其与启动子DNA A-box残基Gua-6的Watson-Crick样H键相互作用的基础上所预测的,在体内用40位的核碱基氨基酸修饰的Rob蛋白表现出改变的DNA启动子结合。Rob蛋白表达最终控制表型变化,包括对抗生素的抗性。尽管带有核碱基氨基酸的Rob蛋白在大肠杆菌中的表达水平估计仅为野生型Rob蛋白的一小部分,但与micF启动子结合的修饰蛋白比野生型在体外的亲和力更高。体内对大环内酯类抗生素罗红霉素和克拉霉素以及β-内酰胺类抗生素氨苄青霉素具有耐药性。还证明了关键的Rob类似物改变的DNA结合和抗生素抗性的统计意义。这些初步发现表明,核碱基氨基酸在通过蛋白质改变和控制优选的核酸靶序列方面具有最终用途,可用于探测对蛋白质功能至关重要的分子相互作用,
更新日期:2020-03-19
中文翻译:

体内转录调节剂Rob的改变:在存在核碱基氨基酸的情况下增强启动子DNA结合和抗生素抗性。
选择性结合核酸序列的蛋白质的鉴定是一个持续的挑战。我们先前合成了旨在取代蛋白原性氨基酸的核碱基氨基酸;将它们掺入蛋白质以可预测地结合特异性核酸。一个较早的例子涉及hnRNP LL RRM1结构域通过Watson-Crick样的H键相互作用选择性地无细胞结合其i-基序DNA靶标。在这项研究中,我们采用了转录调节器Rob的X射线晶体结构,该结构与其micF启动子结合,而DNA不会发生畸变。如在其与启动子DNA A-box残基Gua-6的Watson-Crick样H键相互作用的基础上所预测的,在体内用40位的核碱基氨基酸修饰的Rob蛋白表现出改变的DNA启动子结合。Rob蛋白表达最终控制表型变化,包括对抗生素的抗性。尽管带有核碱基氨基酸的Rob蛋白在大肠杆菌中的表达水平估计仅为野生型Rob蛋白的一小部分,但与micF启动子结合的修饰蛋白比野生型在体外的亲和力更高。体内对大环内酯类抗生素罗红霉素和克拉霉素以及β-内酰胺类抗生素氨苄青霉素具有耐药性。还证明了关键的Rob类似物改变的DNA结合和抗生素抗性的统计意义。这些初步发现表明,核碱基氨基酸在通过蛋白质改变和控制优选的核酸靶序列方面具有最终用途,可用于探测对蛋白质功能至关重要的分子相互作用,


























 京公网安备 11010802027423号
京公网安备 11010802027423号