PLOS Genetics ( IF 4.5 ) Pub Date : 2020-03-09 , DOI: 10.1371/journal.pgen.1008646 Yufeng Fang 1 , Marco A Coelho 1 , Haidong Shu 2 , Klaas Schotanus 1 , Bhagya C Thimmappa 3 , Vikas Yadav 1 , Han Chen 2 , Ewa P Malc 4 , Jeremy Wang 4 , Piotr A Mieczkowski 4 , Brent Kronmiller 5 , Brett M Tyler 5 , Kaustuv Sanyal 3 , Suomeng Dong 2 , Minou Nowrousian 6 , Joseph Heitman 1
|
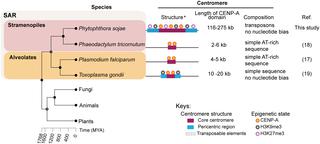
Centromeres are chromosomal regions that serve as platforms for kinetochore assembly and spindle attachments, ensuring accurate chromosome segregation during cell division. Despite functional conservation, centromere DNA sequences are diverse and often repetitive, making them challenging to assemble and identify. Here, we describe centromeres in an oomycete Phytophthora sojae by combining long-read sequencing-based genome assembly and chromatin immunoprecipitation for the centromeric histone CENP-A followed by high-throughput sequencing (ChIP-seq). P. sojae centromeres cluster at a single focus at different life stages and during nuclear division. We report an improved genome assembly of the P. sojae reference strain, which enabled identification of 15 enriched CENP-A binding regions as putative centromeres. By focusing on a subset of these regions, we demonstrate that centromeres in P. sojae are regional, spanning 211 to 356 kb. Most of these regions are transposon-rich, poorly transcribed, and lack the histone modification H3K4me2 but are embedded within regions with the heterochromatin marks H3K9me3 and H3K27me3. Strikingly, we discovered a Copia-like transposon (CoLT) that is highly enriched in the CENP-A chromatin. Similar clustered elements are also found in oomycete relatives of P. sojae, and may be applied as a criterion for prediction of oomycete centromeres. This work reveals a divergence of centromere features in oomycetes as compared to other organisms in the Stramenopila-Alveolata-Rhizaria (SAR) supergroup including diatoms and Plasmodium falciparum that have relatively short and simple regional centromeres. Identification of P. sojae centromeres in turn also advances the genome assembly.
中文翻译:

卵菌中富含转座子的长着丝粒揭示了 Stramenopila-Alveolata-Rhizaria 谱系中着丝粒特征的差异。
着丝粒是染色体区域,用作着丝粒组装和纺锤体附着的平台,确保细胞分裂过程中准确的染色体分离。尽管功能保守,但着丝粒 DNA 序列是多种多样的,而且经常重复,这使得它们难以组装和识别。在这里,我们通过结合基于长读长测序的基因组组装和染色质免疫沉淀对着丝粒组蛋白CENP -A 进行高通量测序 (ChIP-seq) 来描述卵菌中的着丝粒。磷。大豆着丝粒在不同生命阶段和核分裂期间聚集在一个焦点上。我们报告了P的改进基因组组装。酱油参考菌株,它能够将 15 个富集的 CENP-A 结合区域识别为推定的着丝粒。通过关注这些区域的一个子集,我们证明了P中的着丝粒。sojae是区域性的,从 211 到 356 kb。这些区域中的大多数富含转座子,转录不良,缺乏组蛋白修饰 H3K4me2,但嵌入具有异染色质标记 H3K9me3 和 H3K27me3 的区域内。引人注目的是,我们发现了一种在 CENP-A 染色质中高度富集的Copia样转座子 (CoLT)。在P的卵菌近亲中也发现了类似的成簇元素。酱油, 并可用作预测卵菌着丝粒的标准。这项工作揭示了卵菌的着丝粒特征与 Stramenopila-Alveolata-Rhizaria (SAR) 超群中的其他生物相比存在差异,包括硅藻和恶性疟原虫,它们具有相对较短和简单的区域着丝粒。P的识别。大豆着丝粒反过来也促进了基因组组装。


























 京公网安备 11010802027423号
京公网安备 11010802027423号