PLoS Pathogens ( IF 6.7 ) Pub Date : 2020-03-09 , DOI: 10.1371/journal.ppat.1008344 Robert C Shields 1 , Alejandro R Walker 1 , Natalie Maricic 1 , Brinta Chakraborty 1 , Simon A M Underhill 2 , Robert A Burne 1
|
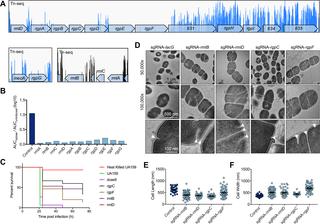
A recent genome-wide screen identified ~300 essential or growth-supporting genes in the dental caries pathogen Streptococcus mutans. To be able to study these genes, we built a CRISPR interference tool around the Cas9 nuclease (Cas9Smu) encoded in the S. mutans UA159 genome. Using a xylose-inducible dead Cas9Smu with a constitutively active single-guide RNA (sgRNA), we observed titratable repression of GFP fluorescence that compared favorably to that of Streptococcus pyogenes dCas9 (Cas9Spy). We then investigated sgRNA specificity and proto-spacer adjacent motif (PAM) requirements. Interference by sgRNAs did not occur with double or triple base-pair mutations, or if single base-pair mutations were in the 3’ end of the sgRNA. Bioinformatic analysis of >450 S. mutans genomes allied with in vivo assays revealed a similar PAM recognition sequence as the Cas9Spy. Next, we created a comprehensive library of sgRNA plasmids that were directed at essential and growth-supporting genes. We discovered growth defects for 77% of the CRISPRi strains expressing sgRNAs. Phenotypes of CRISPRi strains, across several biological pathways, were assessed using fluorescence microscopy. A variety of cell structure anomalies were observed, including segregational instability of the chromosome, enlarged cells, and ovococci-to-rod shape transitions. CRISPRi was also employed to observe how silencing of cell wall glycopolysaccharide biosynthesis (rhamnose-glucose polysaccharide, RGP) affected both cell division and pathogenesis in a wax worm model. The CRISPRi tool and sgRNA library are valuable resources for characterizing essential genes in S. mutans, some of which could prove to be promising therapeutic targets.
中文翻译:

重新改造变形链球菌CRISPR-Cas9系统以了解基本基因功能。
最近的全基因组筛选在龋齿病原体变形链球菌中发现了约300个必需基因或支持生长的基因。为了能够研究这些基因,我们围绕S编码的Cas9核酸酶(Cas9 Smu)构建了一个CRISPR干扰工具。变形UA159基因组。使用木糖诱导的死Cas9 Smu和组成型活性单向导RNA(sgRNA),我们观察到可滴定的GFP荧光抑制,与化脓性链球菌dCas9(Cas9 Spy)。然后,我们研究了sgRNA特异性和原型间隔子相邻基序(PAM)的要求。sgRNA的干扰不会出现双碱基或三碱基对突变,或者单碱基对突变位于sgRNA的3'末端。> 450 S的生物信息学分析。与体内测定相关的变形基因组揭示了与Cas9 Spy类似的PAM识别序列。接下来,我们创建了一个完整的sgRNA质粒文库,该文库针对必需和支持生长的基因。我们发现77%的表达sgRNA的CRISPRi菌株存在生长缺陷。使用荧光显微镜评估了跨多个生物学途径的CRISPRi菌株的表型。观察到各种细胞结构异常,包括染色体的分离不稳定性,扩大的细胞和卵球菌到杆状的形状转变。CRISPRi还用于观察细胞壁糖多糖生物合成的沉默(鼠李糖-葡萄糖多糖,RGP)如何影响蜡虫模型中的细胞分裂和发病机理。CRISPRi工具和sgRNA文库是鉴定S必需基因的宝贵资源。变种人,其中一些可能被证明是有希望的治疗靶标。


























 京公网安备 11010802027423号
京公网安备 11010802027423号