当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Harnessing β-Lactam Antibiotics for Illumination of the Activity of Penicillin-Binding Proteins in Bacillus subtilis.
ACS Chemical Biology ( IF 4 ) Pub Date : 2020-03-20 , DOI: 10.1021/acschembio.9b00977 Shabnam Sharifzadeh , Felix Dempwolff 1 , Daniel B Kearns 1 , Erin E Carlson
ACS Chemical Biology ( IF 4 ) Pub Date : 2020-03-20 , DOI: 10.1021/acschembio.9b00977 Shabnam Sharifzadeh , Felix Dempwolff 1 , Daniel B Kearns 1 , Erin E Carlson
Affiliation
|
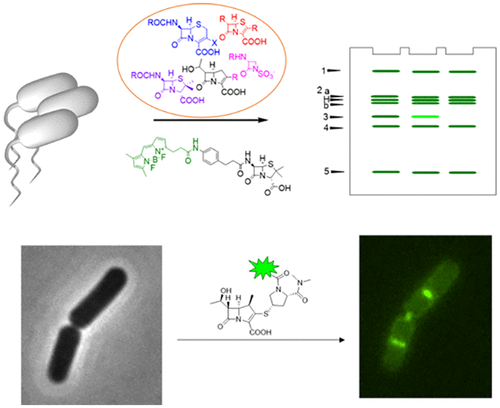
Selective chemical probes enable individual investigation of penicillin-binding proteins (PBPs) and provide critical information about their enzymatic activity with spatial and temporal resolution. To identify scaffolds for novel probes to study peptidoglycan biosynthesis in Bacillus subtilis, we evaluated the PBP inhibition profiles of 21 β-lactam antibiotics from different structural subclasses using a fluorescence-based assay. Most compounds readily labeled PBP1, PBP2a, PBP2b, or PBP4. Almost all penicillin scaffolds were coselective for all or combinations of PBP2a, 2b, and 4. Cephalosporins, on the other hand, possessed the lowest IC50 values for PBP1 alone or along with PBP4 (ceftriaxone, cefoxitin) and 2b (cefotaxime) or 2a, 2b, and 4 (cephalothin). Overall, five selective inhibitors for PBP1 (aztreonam, faropenem, piperacillin, cefuroxime, and cefsulodin), one selective inhibitor for PBP5 (6-aminopenicillanic acid), and various coselective inhibitors for other PBPs in B. subtilis were discovered. Surprisingly, carbapenems strongly inhibited PBP3, formerly shown to have low affinity for β-lactams and speculated to be involved in β-lactam resistance in B. subtilis. To investigate the specific roles of PBP3, we developed activity-based probes based on the meropenem core and utilized them to monitor the activity of PBP3 in living cells. We showed that PBP3 activity localizes as patches in single cells and concentrates as a ring at the septum and the division site during the cell growth cycle. Our activity-based approach enabled spatial resolution of the transpeptidation activity of individual PBPs in this model microorganism, which was not possible with previous chemical and biological approaches.
中文翻译:

利用β-内酰胺抗生素来阐明枯草芽孢杆菌中青霉素结合蛋白的活性。
选择性化学探针可对青霉素结合蛋白(PBP)进行单独研究,并提供有关其酶促活性的关键信息(时空分辨率)。为了鉴定用于研究枯草芽孢杆菌中肽聚糖生物合成的新型探针的支架,我们使用基于荧光的分析方法评估了来自不同结构亚类的21种β-内酰胺类抗生素的PBP抑制谱。大多数化合物容易标记为PBP1,PBP2a,PBP2b或PBP4。几乎所有的青霉素支架对PBP2a,2b和4的全部或组合都是共选择性的。另一方面,头孢菌素单独或与PBP4(头孢曲松,头孢西丁)和2b(头孢噻肟)或2a一起具有最低的IC50值, 2b和4(头孢菌素)。总体而言,五种PBP1选择性抑制剂(氨曲南,法罗培南,哌拉西林,头孢呋辛,和头孢磺啶),枯草芽孢杆菌中发现了一种PBP5(6-氨基青霉酸)的选择性抑制剂和其他PBPs的多种共选择性抑制剂。令人惊讶地,碳青霉烯类强烈抑制PBP3,PBP3以前显示对β-内酰胺的亲和力低,并推测与枯草芽孢杆菌的β-内酰胺抗性有关。为了研究PBP3的特定作用,我们开发了基于美洛培南核心的基于活性的探针,并利用它们来监测活细胞中PBP3的活性。我们显示,PBP3活性在单个细胞中定位为斑块,并在细胞生长周期中集中在隔片和分裂部位的环上。我们基于活动的方法能够对这种模型微生物中单个PBP的转肽活性进行空间解析,
更新日期:2020-03-10
中文翻译:

利用β-内酰胺抗生素来阐明枯草芽孢杆菌中青霉素结合蛋白的活性。
选择性化学探针可对青霉素结合蛋白(PBP)进行单独研究,并提供有关其酶促活性的关键信息(时空分辨率)。为了鉴定用于研究枯草芽孢杆菌中肽聚糖生物合成的新型探针的支架,我们使用基于荧光的分析方法评估了来自不同结构亚类的21种β-内酰胺类抗生素的PBP抑制谱。大多数化合物容易标记为PBP1,PBP2a,PBP2b或PBP4。几乎所有的青霉素支架对PBP2a,2b和4的全部或组合都是共选择性的。另一方面,头孢菌素单独或与PBP4(头孢曲松,头孢西丁)和2b(头孢噻肟)或2a一起具有最低的IC50值, 2b和4(头孢菌素)。总体而言,五种PBP1选择性抑制剂(氨曲南,法罗培南,哌拉西林,头孢呋辛,和头孢磺啶),枯草芽孢杆菌中发现了一种PBP5(6-氨基青霉酸)的选择性抑制剂和其他PBPs的多种共选择性抑制剂。令人惊讶地,碳青霉烯类强烈抑制PBP3,PBP3以前显示对β-内酰胺的亲和力低,并推测与枯草芽孢杆菌的β-内酰胺抗性有关。为了研究PBP3的特定作用,我们开发了基于美洛培南核心的基于活性的探针,并利用它们来监测活细胞中PBP3的活性。我们显示,PBP3活性在单个细胞中定位为斑块,并在细胞生长周期中集中在隔片和分裂部位的环上。我们基于活动的方法能够对这种模型微生物中单个PBP的转肽活性进行空间解析,



























 京公网安备 11010802027423号
京公网安备 11010802027423号