Journal of Structural Biology ( IF 3 ) Pub Date : 2020-02-17 , DOI: 10.1016/j.jsb.2020.107479 K Muthuvel Prasath 1 , K Ganesan 2 , S Parthasarathy 1
|
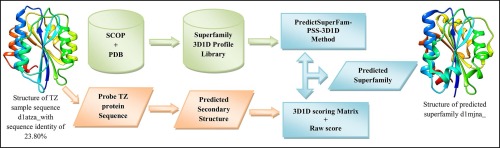
Annotation of twilight zone protein sequences has been hitherto attempted by predicting the fold of the given sequence. We report here the PredictSuperFam-PSS-3D1D method, which predicts the superfamily for a given twilight zone (TZ) protein sequence. Earlier, we have reported that adding predicted secondary structure information into the threading methods could improve fold prediction especially for the TZ protein sequences. In this study, we have analysed the application of the same method to predict superfamilies. Here, in this method, the twilight zone protein sequence is threaded with the 3D1D profiles of the known protein superfamilies library. In addition, weightage for the predicted secondary structure (PSS) is also employed. The performance of the method is benchmarked with twilight zone sequences. In the benchmarks, 62 and 65 percentages of superfamily predictions are obtained with GOR IV and NPS@ predicted secondary structures, respectively. Receiver Operating Characteristic (ROC) curves indicate that the method is sensitive in predicting the superfamilies. A case study has been conducted with the hypothetical protein sequences of Schistosoma haematobium (Blood Fluke) using this method and the results are analyzed. Our method predicts the superfamily for TZ sequences for which, methods based on sequence similarity alone are inadequate. A web server has been developed for our method and it is available online at http://bioinfo.bdu.ac.in/psfpss.
中文翻译:

PredictSuperFam-PSS-3D1D:用于预测超家族的服务器,用于注释暮光区蛋白序列。
迄今为止,已经通过预测给定序列的折叠来尝试对暮光区蛋白序列进行注释。我们在这里报告PredictSuperFam-PSS-3D1D方法,该方法预测给定暮光区(TZ)蛋白序列的超家族。此前,我们已经报道了将预测的二级结构信息添加到穿线方法中可以改善折叠预测,尤其是对于TZ蛋白序列。在这项研究中,我们分析了相同方法在预测超家族中的应用。在这里,在这种方法中,暮光区蛋白质序列与已知蛋白质超家族文库的3D1D谱图连接在一起。此外,预测二级结构的权重(PSS)也可以使用。该方法的性能以暮光区序列为基准。在基准测试中,通过GOR IV和NPS @预测的二级结构分别获得了62%和65%的超家族预测。接收者操作特征(ROC)曲线表明该方法在预测超家族方面很敏感。使用这种方法对血吸虫血吸虫假想的蛋白质序列进行了案例研究,并对结果进行了分析。我们的方法预测了TZ序列的超家族,对于这些家族而言,仅基于序列相似性的方法是不够的。已经为我们的方法开发了一个Web服务器,可以从http://bioinfo.bdu.ac.in/psfpss在线获取。


























 京公网安备 11010802027423号
京公网安备 11010802027423号